Nikhil Prasad Fact checked by:Thailand Medical News Team May 29, 2024 10 months, 4 weeks, 16 hours, 44 minutes ago
COVID-19 News: In an unprecedented study, researchers from Augusta University, the University of South Carolina, and Greenwood Genetic Center have shed new light on the complex immune dysregulation caused by SARS-CoV-2. This comprehensive analysis of post-mortem lung tissues from COVID-19 victims covered in this
COVID-19 News report, reveals significant alterations in gene expression, providing crucial insights into the disease's pathogenesis and potential therapeutic targets.
 Autopsy Study Validates That SARS-CoV-2 Causes Immune Dysregulation
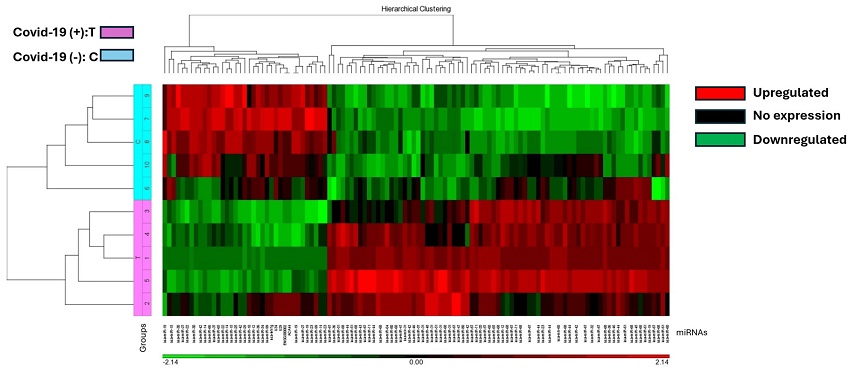
The heat map illustrates miRNA expression patterns in COVID-19 positive (T) and negative (C) lung biopsies. Five samples from each group were analyzed using the Affymetrix array system. miRNAs are represented as columns, while samples are depicted as rows. Expression levels are color-coded: red indicates higher expression, black indicates no expression, and green indicates lower expression. COVID-19 positive samples are denoted in pink, while negative samples are highlighted in blue.
The Immune System's Role in COVID-19 Severity
Autopsy Study Validates That SARS-CoV-2 Causes Immune Dysregulation
The heat map illustrates miRNA expression patterns in COVID-19 positive (T) and negative (C) lung biopsies. Five samples from each group were analyzed using the Affymetrix array system. miRNAs are represented as columns, while samples are depicted as rows. Expression levels are color-coded: red indicates higher expression, black indicates no expression, and green indicates lower expression. COVID-19 positive samples are denoted in pink, while negative samples are highlighted in blue.
The Immune System's Role in COVID-19 Severity
COVID-19, caused by the novel coronavirus SARS-CoV-2, has emerged as one of the most significant health crises in recent history. The clinical characteristics of COVID-19 patients suggest that changes in immune activity play a critical role in disease severity. However, detailed information about the immune response in human lung tissue, the primary site of infection, remains limited.
To address this gap, the research team conducted an extensive analysis of lung tissues from five individuals who died from COVID-19 and seven control individuals who died from unrelated causes. The study aimed to identify differentially expressed miRNAs and mRNAs, providing a clearer understanding of the host response to the virus.
Methodology: Microarray and Gene Expression Assays
To analyze the host response, the researchers used miRNA microarray and Nanostring’s nCounter XT gene expression assay. These advanced techniques allowed the identification of 37 downregulated and 77 upregulated miRNAs in COVID-19 lung biopsy samples compared to controls. Additionally, 653 mRNA transcripts were differentially expressed, with the majority (472) being downregulated in COVID-19-positive specimens.
Distinct Clustering of COVID-19 and Control Samples
Hierarchical and Principal Component Analysis (PCA) K-means clustering revealed distinct differences between COVID-19 and control samples. Enrichment and network analyses highlighted the importance of genes involved in innate immunity and inflammatory response in COVID-19 lung biopsies. Notably, the interferon-signaling pathway was highly upregulated in COVID-19 specimens, while genes involved in interleukin-17 signaling were downregulated.
The Lungs: A Central Organ in COVID-19 Pathology
The lung is a primary site of infection and organ failure in COVID-19. The virus spreads through respiratory droplets and binds to ACE2 receptors in the nasal epithelium, facilitating transmission to the lower respiratory tract. After initial infection, the host response progresses through stages marked by macrophages, dendritic cells, and cytotoxic T cells. Failure to clear the infection can lead to a hyperinflammatory stage, cytokine storm, and ultimately, multi-organ failure.
Molecular Insights into Lung Tissue Response
Understanding the molecular processes in the lungs during severe COVID-19 infections is crucial for developing life-saving therapies. Despite this, only a limited number of studies have investigated the molecular mechanisms in clinical lung tissues. Previous research indicates that SARS-CoV-2 can repress the type I interferon (IFN) response, a critical antiviral defense mechanism.
Combined Evaluation of mRNA and miRNA Expression
This study represents one of the earliest attempts to simultaneously analyze miRNA and mRNA in COVID-19 patients at an organ level. The researchers aimed to explore transcriptome profiles in patients who succumbed to severe COVID-19 compared to those who died from unrelated causes. The investigation revealed distinct differences in miRNA and mRNA expression patterns between post-mortem lung biopsy specimens of COVID-19-positive and negative individuals.
Key Findings: miRNA and mRNA Expression Alterations
The study identified significant alterations in miRNA and mRNA expression in COVID-19-infected lung tissues. A total of 37 miRNAs were downregulated and 77 miRNAs were upregulated in COVID-19-positive lung biopsy specimens.
Hierarchical cluster analysis generated a heatmap of differentially expressed miRNAs, revealing a distinct split between COVID-19 and control cases.
Similarly, a total of 653 mRNA transcripts were differentially expressed, with 472 downregulated and 170 upregulated in COVID-19-positive specimens. The mRNA heatmap generated using Nanostring Advanced Analysis software showed distinct clustering between COVID-19-positive and negative patients.
Differential Gene Expression and Pathway Enrichment
Further analysis revealed that genes involved in innate immunity were highly differentially expressed in COVID-19-positive lung biopsy specimens. The interferon-signaling pathway, particularly type I and type III interferon responses, were significantly upregulated. Conversely, pathways related to tissue stress, ALPK1 signaling, and leukotriene and prostaglandin inflammation were downregulated.
Immune Dysregulation in Severe COVID-19 Infection
The study highlights immune dysregulation as a crucial factor in the progression of severe COVID-19 infection. Specifically, genes involved in neutrophil recruitment and chronic inflammation, such as CXCL5, CXCL8, and IL-6, were significantly downregulated in COVID-19-positive tissues. This suggests that immune dysregulation plays a critical role in COVID-19 pathogenesis.
The Role of Innate Immunity and Toll-Like Receptors
The innate immune system is vital for detecting and responding to infections. In COVID-19, Toll-like receptors (TLRs), especially TLR7 and TLR8, which bind to single-stranded RNA, are enriched in lung tissues and play critical roles in the immune response. Activation of TLR pathways leads to the production of pro-inflammatory cytokines and type I interferon (IFN-1), crucial for antiviral defense.
The ISG15 Pathway and Its Implications
The ISG15 pathway, involved in the antiviral response, was significantly upregulated in COVID-19-positive lung tissues. Genes such as ISG15, IFIT1, RSAD2, OAS1, MX1, and IFIT3 were highly expressed. While the ISG15 pathway is essential for antiviral defense, its role in promoting local inflammation and poor outcomes in COVID-19 is emerging as a critical area of research.
miRNAs and Their Regulatory Role
Several miRNAs known to repress the ISG15 pathway were identified as downregulated in COVID-19-positive tissues. For example, hsa-miR-146a-5p, a miRNA that modulates inflammation, was significantly downregulated. This miRNA targets genes involved in the ISG15 pathway, highlighting its regulatory role in the antiviral response and inflammation.
Th1 and Th17 Responses in COVID-19
Functional enrichment analysis revealed a trend towards upregulation of Th1 response genes and downregulation of Th17 response genes in COVID-19-positive lung tissues. This finding aligns with the current understanding of immunology, suggesting that Th1-mediated responses play a more significant role in severe COVID-19 lung infections.
Th2-Mediated Immune Responses
Interestingly, Th2-supporting cytokines, such as IL5 and IL13, were among the most positively upregulated mRNAs in COVID-19-positive samples. Th2 cell-mediated immune functions are generally associated with allergic and atopic responses. The study's findings suggest that Th2-mediated allergic reactions could be present in severe COVID-19 infections, potentially contributing to disease severity.
Implications for Treatment and Prevention
The study's findings highlight the importance of understanding immune dysregulation in COVID-19 pathogenesis. The differential expression of genes involved in immune regulation suggests potential therapeutic targets for mitigating the effects of severe COVID-19 infection. Further research is needed to explore the regulatory roles of miRNAs and the modulation of immune responses in COVID-19.
Conclusion
This groundbreaking study validates that SARS-CoV-2 causes significant immune dysregulation, particularly in lung tissues. By analyzing miRNA and mRNA expression patterns, the researchers have provided valuable insights into the molecular mechanisms underlying severe COVID-19 infection. These findings pave the way for the development of targeted therapies to address immune dysregulation and improve outcomes for COVID-19 patients.
The study findings were published in the peer reviewed journal: Viruses.
https://www.mdpi.com/1999-4915/16/6/853
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
Read Also:
https://www.thailandmedical.news/news/sars-cov-2-e-protein-induces-long-term-immune-dysfunction
https://www.thailandmedical.news/news/type-i-interferon-autoantibodies-in-covid-19-cause-immune-dysregulation-and-contribute-to-disease-severity
