BREAKING! ACE2 Has Novel Protein Interactions That Are Disrupted During SARS-CoV-2 Infection, Contributing To The Spectrum Of COVID-19 Pathologies!
Source: SARS-CoV-2 Research Oct 01, 2021 3 years, 6 months, 3 weeks, 4 days, 5 hours, 26 minutes ago
SARS-CoV-2 Research: A new study by researchers from University of Rochester has uncovered that the receptors ACE2 has novel protein interactions that are disrupted during SARS-CoV-2 infection, contributing to the spectrum of COVID-19 pathologies!
.jpg)
The ACE2 or angiotensin-converting enzyme 2 is the cell receptor that the SARS-CoV-2 coronavirus binds to and uses to enter and infect human cells.
The COVID-19 disease caused by the novel coronavirus, involves diverse pathologies beyond those of a respiratory disease, including micro-thrombosis (micro-clotting), cytokine storms, and inflammatory responses affecting many organ systems.
Longer-term chronic illness can persist for many months, often well after the pathogen is no longer detected. A better understanding of the proteins that ACE2 interacts with can reveal information relevant to these disease manifestations and possible avenues for treatment.
The
SARS-CoV-2 Research team had undertaken an approach to predict candidate ACE2 interacting proteins which uses evolutionary inference to identify a set of mammalian proteins that “coevolve” with ACE2.
This new approach, called evolutionary rate correlation (ERC), detects proteins that show highly correlated evolutionary rates during mammalian evolution.
Such identified proteins are candidates for biological interactions with the ACE2 receptor.
Importantly the approach has uncovered a number of key ACE2 protein interactions of potential relevance to COVID-19 pathologies. Some proteins have previously been reported to be associated with severe COVID-19, but are not currently known to interact with ACE2, while additional predicted novel ACE2 interactors are of potential relevance to the disease.
Utilizing reciprocal rankings of protein ERCs, the study team identified strongly interconnected ACE2 associated protein networks relevant to COVID-19 pathologies.
The study findings showed that ACE2 has clear connections to coagulation pathway proteins, such as Coagulation Factor V and fibrinogen components FGA, FGB, and FGG, the latter possibly mediated through ACE2 connections to Clusterin (which clears misfolded extracellular proteins) and GPR141 (whose functions are relatively unknown).
The study findings also showed that ACE2 connects to proteins involved in cytokine signaling and immune response (e.g. XCR1, IFNAR2 and TLR8), and to Androgen Receptor (AR).
The ERC prescreening approach has elucidated possible functions for relatively uncharacterized proteins and possible new functions for well-characterized ones. Suggestions are made for the validation of ERC-predicted ACE2 protein interactions.
The study findings conclude that ACE2 has novel protein interactions that are disrupted during SARS-CoV-2 infection, contributing to the wide spectrum of COVID-19 pathologies.
The study findings were published in the peer reviewed journal: Bioinformatics and Genomics. (Peer J Life and Environment)
https://peerj.com/articles/12159/<
br />
The COVID-19 disease not only causes symptoms characteristic of a typical respiratory disorder, but has also been known to trigger a wide range of other symptoms in individuals who had been infected, some lasting even long after individuals test negative for the virus. These symptoms can include abnormal blood clotting, heart damage and failure, kidney disease, brain fog (confusion, memory loss, or difficulty focusing), gastrointestinal problems, and even male infertility.
However to date, the mechanisms by which COVID-19 causes these diverse complications remain poorly understood.
The study team led by Dr John H Werren, the Nathaniel and Helen Wisch Professor of Biology at the University of Rochester, and recent undergraduates Austin Varela '20 and Sammy Cheng '21 studied proteins that closely evolve with angiotensin-converting enzyme 2 (ACE2), the receptor used by the SARS-CoV2coronavirus to enter human cells.
Utilizing an evolutionary approach, the study team detected proteins that ‘coevolve’ with ACE2 in mammals as a way to identify networks of proteins that likely interact with ACE2 during its normal functions in the human body.
The study team came to the conclusion that disruptions caused by COVID-19 in these normal functions of ACE2 could contribute to the unusual pathologies of the disease.
The study findings revealed a number of candidate protein partners for ACE2 that have not previously been identified as ACE2 interactors, but which could have direct bearing on the complications experienced by individuals infected with the virus.
Interestingly these COVID-related complications can include excessive blood clotting as well as an overactive inflammation response known as a ‘cytokine storm’, both of which can cause tissue and organ damage.
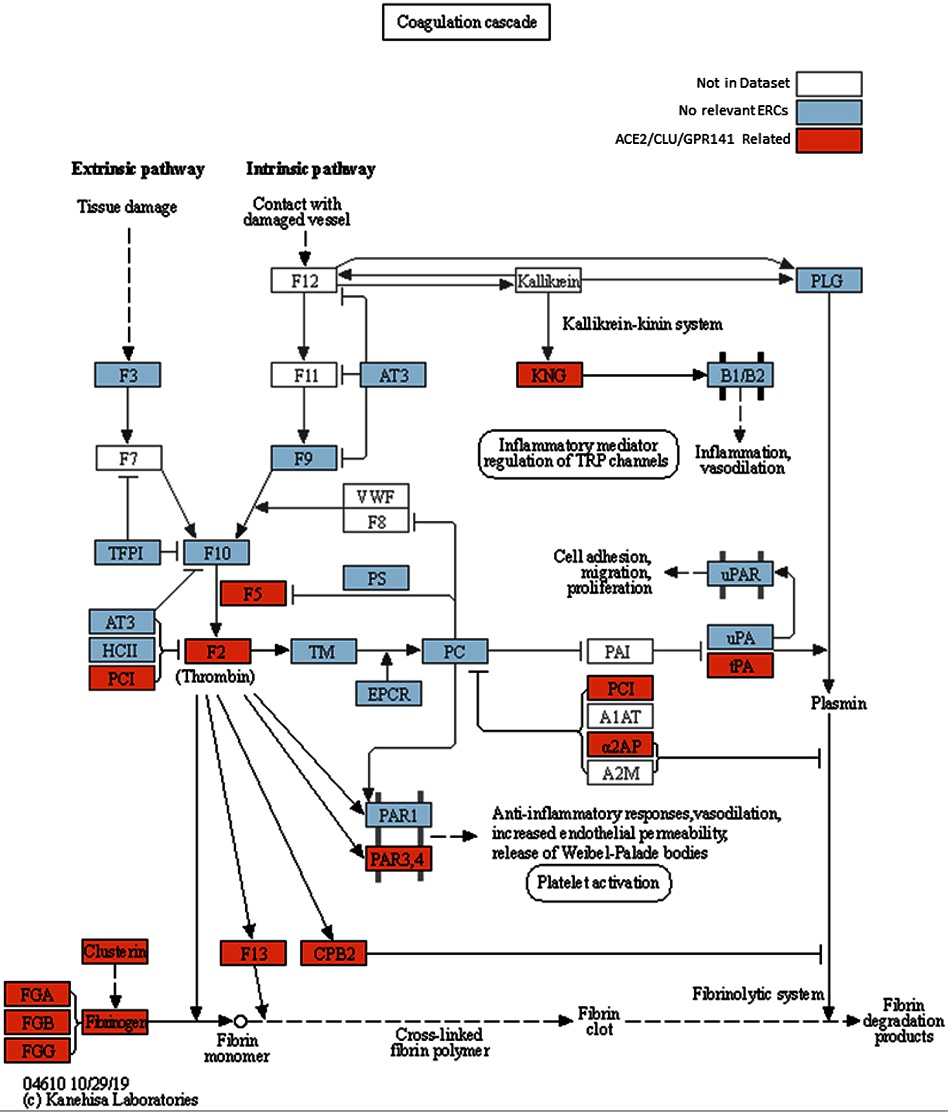 Modified KEGG coagulation pathway.
Modified KEGG coagulation pathway.
KEGG Coagulation cascade pathway, with ACE2-CLU-GPR141 associated proteins (based on presence in any of their top 2% ERCs or in the ACE2 CRR network) indicated in orange. The KEGG pathway has been supplemented to indicate the three fibrinogen proteins and clusterin associations previously discussed. Note the alternate protein names: PAR3,4 = F2RL2 & F2RL3 = Thrombin receptors; α2AP = Alpha-2-antiplasmin = SERPINF2; PLAT = tPA, and PCI = SERPINA5 = Protein C Inhibitor.
As an example, one hallmark of severe COVID-19 is abnormal blood coagulation throughout the body. The study team's research revealed noteworthy connections between ACE2 and key proteins involved in the coagulation pathway. Another protein, Clusterin, which plays a significant role in "quality control" in the blood by removing misfolded proteins, strongly coevolves with ACE2-;implying that they interact with each other biologically. Several proteins involved in cytokine signaling appear to coevolve with ACE2 as well.
Dr Werren told Thailand Medical News, "We propose that ACE2 has novel protein interactions that are disrupted during SARS-CoV-2 infection, contributing to the spectrum of COVID-19 pathologies.”
.jpg) ACE2 centric reciprocal rank (CRR) network.
ACE2 centric reciprocal rank (CRR) network.
Proteins with ERC reciprocal ranks ≤20 are shown by double-headed arrows, and unidirectional ranks ≤20 connecting to the RR backbone are indicated by single-headed arrows. ACE2 has extensive connections to coagulation proteins mediated primarily through Clusterin (CLU) and GPR141. ACE2 is highlighted in purple, and blue shading intensity indicates the level of reciprocal connectivity for different proteins.
Importantly finding that ACE2's evolutionary partners are involved in blood coagulation and cytokine signaling is consistent with this possibility.
Dr Werren further added, “However these candidate protein interactions will need to be validated. But if supported, the study findings could inform development of better treatments and therapeutics for COVID-19 and chronic complications that may arise."
Significantly as an evolutionary geneticist, Dr Werren's research focuses on the interaction of genomes in symbiotic or parasitic relationships. When the COVID-19 pandemic hit, Dr Werren received a grant from the National Science Foundation's Rapid Response Research program to study the ACE2's protein interactions and its network of coevolving proteins. The University's Nathaniel and Helen Wisch Chair Research Fund, meanwhile, helped support Varela's and Cheng's participation.
Varela, the study's first author, who started researching protein-protein interactions in the Werren Lab during his sophomore year at Rochester commented, "Working on this project was a great opportunity for me.Once we uncovered the evolutionary rate partners of ACE2, their potential clinical relevance was immediately clear."
In the evolutionary approach to detecting protein partners and interactions
Dr Werren and his colleagues used a computational and evolutionary approach called evolutionary rate correlation (ERC).
The key underlying concept is that proteins with similar rates of change during evolution are more likely to have functional interactions. For example, if you look at a phylogenetic tree depicting the evolutionary relationships among mammals, when one protein evolves quickly in a particular species, a protein with which it functionally interacts will tend to evolve quickly as well, and vice versa.
Dr Werren previously applied the ERC method to protein interactions involved in mitochondrial function. Mitochondria are cellular structures that, among other functions, produce energy for the cell. In that study, the ERC method accurately predicted nuclear encoded proteins known to interact with mitochondria, and also detected proteins not previously known to have mitochondrial function.
Bat present biomedical researchers are able to harness a number of powerful tools for detecting and exploring proteins that interact with each other in biological processes. These include genetic screens, protein coprecipitation, and proteomic profiling.
Such evolutionary approaches such as ERC have the potential to become useful additions to the biomedical research toolkit.
Dr Werren and his team are now expanding the number of proteins in their analysis.
Dr Werren added, "We hope to produce a more extensive database for researchers so they can explore coevolving partners for proteins of interest in their research."
The study team concluded, “Our key findings are that ACE2 has previously unidentified protein partners that are part of various interaction networks relevant to COVID-19 pathologies. Most notably, ACE2 forms strong ERC networks relevant to coagulation and immunity. A potential mechanism is that reduced abundance of membrane-bound ACE2 disrupts these signaling networks. Additionally, the presence of the soluble ACE2 ectodomain may explain the systemic pathologies of COVID-19 infection as its circulation in the blood can affect pathways throughout the body. We recognize that the new ACE2 protein connections predicted by ERCs may not be causal in severe COVID-19 pathologies. However, our novel findings that the ACE2 ERC network connects to coagulation and immunity pathways is noteworthy, with clear potential implications to some of the unusual features of COVID-19. In addition, results may have relevance to other functions of ACE2, such as circulatory homeostasis and digestion. The ERC analysis predicts additional protein connections that can be relevant to biological processes and disease. For instance, ERCs predict novel interactions for cytokine and immunity related proteins, such as for XCR1, IFNLR1, IFNAR2, and TLR8. Future investigations of the ERC networks of these and related proteins could be worthwhile. ERCs also suggest strong but previously undescribed connections for proteins such as CLU, GPR141, F5, and GEN1. Validation studies are necessary to determine to what extent strong ERCs predict biological interactions among proteins, such as the ones detected here.”
For the latest
SARS-CoV-2 Research, keep on logging to Thailand Medical News.
.jpg)

.jpg)