BREAKING! COVID-19 Genomics: SARS-CoV-2 Has Mechanism To Mimic Host Cellular mRNAs, Protecting Itself From Host Innate Immune Restriction
Source: COVID-19 Genomics Jul 24, 2020 4 years, 8 months, 3 weeks, 4 days, 20 hours, 39 minutes ago
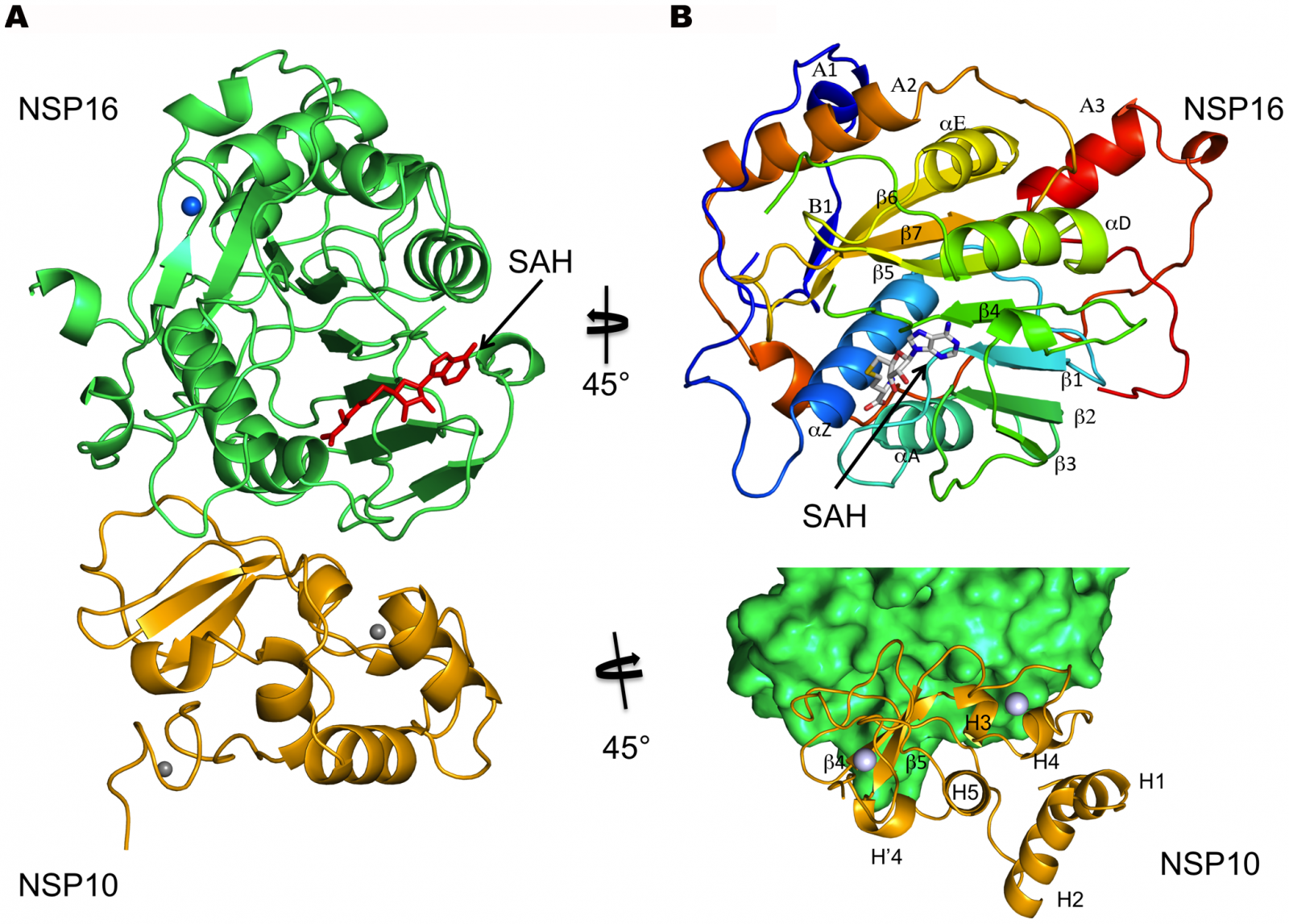
COVID-19 Genomics: Researchers from the University of Texas Health Science Center at San Antonio (UT Health San Antonio) have discovered how the non-structural protein 16 (nsp16) of the SARS-CoV-2 , in conjunction with nsp10, methylates the 5′-end of virally encoded mRNAs to mimic cellular mRNAs, thus protecting the virus from host innate immune restriction.
The research findings were published in the journal: Nature Communications.
https://www.nature.com/articles/s41467-020-17496-8
The researchers resolved the structure of an enzyme called nsp16, which the virus produces and then uses to modify its messenger RNA cap, said Dr Yogesh Gupta, Ph.D., the study lead author from the Joe R. and Teresa Lozano Long School of Medicine at UT Health San Antonio.
The researchers report the high-resolution structure of a ternary complex of SARS-CoV-2 nsp16 and nsp10 in the presence of cognate RNA substrate analogue and methyl donor, S-adenosyl methionine (SAM). The nsp16/nsp10 heterodimer was captured in the act of 2′-O methylation of the ribose sugar of the first nucleotide of SARS-CoV-2 mRNA. The researchers said that they observed large conformational changes associated with substrate binding as the enzyme transitions from a binary to a ternary state. This induced fit model provides mechanistic insights into the 2′-O methylation of the viral mRNA cap in order to gain entry into the host cell undetected.
Dr Gupta said "It's a camouflage. Because of the modifications, which fool the cell, the resulting viral messenger RNA is now considered as part of the cell's own code and not foreign."
Dr Gupta said that deciphering the 3-D structure of nsp16 paves the way for rational design of antiviral drugs for COVID-19 and other emerging coronavirus infectionsThe drugs, new small molecules, would inhibit nsp16 from making the modifications. The immune system would then pounce on the invading virus, recognizing it as foreign.
Study coauthor Dr Robert Hromas, MD, Professor and dean of the Long School of Medicine said, "Dr Yogesh's work discovered the 3-D structure of a key enzyme of the COVID-19 virus required for its replication and found a pocket in it that can be targeted to inhibit that enzyme. This is a fundamental advance in our understanding of the virus."
Dr Gupta is an assistant professor in the Department of Biochemistry and Structural Biology at UT Health San Antonio and is a member of the university's Greehey Children's Cancer Research Institute.
In lay terms, messenger RNA can be described as a deliverer of genetic code to worksites that produce proteins.
For more on
COVID-19 Genomics, keep on logging to Thailand Medical News.
