BREAKING! COVID-19 News: French Scientists Uncover How SARS-CoV-2 ORF8 Protein Causes Dysregulation Of Gene Expression In Infected Cells
COVID-19 News - SARS-CoV-2 ORF8 Protein Causes Dysregulation Of Gene Expression Dec 20, 2022 2 years, 4 months, 5 hours, 51 minutes ago
COVID-19 News: French researchers from Université de Lorraine, Université Côte d’Azur, Institut de Chimie de Nice, Institut Universitaire de France, and Université Paris Cité have in a new study uncovered the detailed mechanism by which the ORF8 protein of the SARS-CoV-2 causes alterations to the gene expression in infected cells.

It is already known that non-structural accessory proteins in viruses play a key role in hijacking the basic cellular mechanisms, which is essential to promote the virus survival and evasion of the immune system.
The small water-soluble immonuglobulin-like open reading frame 8 (ORF8) protein expressed by SARS-CoV-2 accumulates in the nucleus and may influence the regulation of the gene expression in infected cells.
A past study published in Thailand Medical News’ past
COVID-19 News coverages, by researchers from University of Pennsylvania found that the SARS-CoV-2 virus via the ORF8 protein, uses histone mimicry to disrupt host epigenetic regulation in order to disarm host immune responses.
https://www.thailandmedical.news/news/university-of-pennsylvania-study-finds-that-sars-cov-2-uses-histone-mimicry-to-disrupt-host-epigenetic-regulation-for-disarming-host-immune-responses
The French study team further contributed details to this mechanism by using micro-second time-scale all-atom molecular dynamics simulations.
The French researchers unraveled the structural bases behind the epigenetic action of ORF8.
The study team showed how the protein is able to form stable aggregates with DNA through a histone tail-like motif, and how this interaction is influenced by post-translational modifications, such as acetylation and methylation, which are known epigenetic markers in histones.
The study finding not only details the molecular mechanisms behind the perturbation of the epigenetic regulation caused by the viral infection, but also offers an unusual perspective which may foster the development of more effective antivirals.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2022.12.18.520908v1
The study team uncovering the detailed mechanism by which the SARS-CoV-2 ORF8 protein causes dysfunctional gene expression or what is sometimes also called as epigenetic changes by utilizing a multiscale approach combining protein/nucleic acid docking and µs-scale all atom-molecular dynamic (MD) simulations to present the first model of the complex between ORF8 and double-helical DNA. The obtained stable and persistent aggregates are coherent with the observed coprecipitation of ORF8 with chromatin.
The study team also provided evidence how PTMs (post-translational modifications) modify
the interaction network between DNA and ORF8 and hence are susceptible to induce chromatin transitions.
ORF8 crystallizes in the form of a dimer, while the supra molecular organization may be essential for the Ig-like action, the monomer form is also considered as functional, especially for its interaction with DNA and chromatin.
Of particular importance is the presence, on one of the flexible loops, of the ARKS sequence (residue 35 to 38) and to a lesser extent of the subsequent AP motif (residue 39 and 40). Indeed, this motif presents a very strong sequence similarity with the one found in the flexible tail of the histone 3 (H3). Besides, lysine is also considered a hot-spot for PTMs, which may lead to different outcomes relative to gene expression or silencing. Because of the sequence similarity, and also considering the high density of positive charges, the ARKS sequence may clearly be regarded as one of the leading factors favoring the interaction with DNA, and thus to a more global level chromatin compaction.
The ARKS motif is highly exposed in the monomer, while is relatively buried at the protein/protein interface in the case of the dimer, and hence less accessible for DNA complexation. This fact, also confirms the hypothesis that the monomeric form of ORF8 should be active towards DNA.
In order to confirm the possible formation of stable aggregates between ORF8 and DNA, the study team performed protein/nucleic acid docking to obtain starting conformations. Two poses have emerged from the procedure as the most stable or mostly populated cluster.
The two poses have further been relaxed through µs-scale all atom MD simulations. The MD has confirmed that ORF8 is interacting persistently with DNA strands.
However, the first docking pose, which already provided DNA contacts with the ARKS motif evolves only slightly and leads to the insertion of the ORF8 tail in the DNA major groove. Instead, the second pose, initially presented the ARKS tail oriented away from the duplex. However, upon MD and on the hundreds of ns timescale ORF8 relaxes and slightly slides along the duplex, to finally stabilizing after establishing a contact between ARKS and the nucleic acid minor groove.
Although globally stable, the two interaction modes give rise to slightly different interaction patterns involving, mainly, the nucleic acid backbone. Notably, however, both the major and the minor groove bound states involve the ORF8 RKS sequence which develops rather persistent interaction with the nucleic acid fragments.
The study findings prove that ORF8 is indeed able to stably bind DNA via its histone-mimicry ARKS motif. More specifically, two motifs have been evidenced involving either the DNA major or minor groove Whether, the second one is probably less favorable, both motifs may be competitive to assure DNA binding. Therefore, ORF8 is susceptible of influencing the compaction of the chromatin and hence to act as a perturbator of the epigenetic regulations of the host cells.
The ARKS sequence in the H3 histone being also a hotspot for PTMs, and particularly lysine acetylation and methylation.
The study team examined their effects on the stability of the ORF8/DNA aggregate and on the modification of the established interaction network. Even upon the presence of PTMS, and more specifically lysine acetylation or methylation, the ORF8/DNA complex remained stable throughout the MD simulation, without showing any relevant structural modification either in both interaction modes.
Unsurprisingly, however, the interaction of K37 with the DNA backbone is lost, whatever the PTM considered. This loss of interaction, which could involve chromatin remodeling, and favor gene expression, is however not accompanied by a noticeable weakening of the contacts experienced by the ARKS motif with DNA. Indeed, both the interaction of R36 and S38 are highly conserved.
Only rather negligible differences in the distribution of the distances compared to the native protein are evidenced in both the position and the shape of the main peak. Globally a more pronounced destabilization of the interaction network can be observed for the minor-groove bound mode, which is particularly evident for the acetylated and single-methylated ORF8.
Notably, in the latter case the more important perturbation are observed in the case of acetylation or single methylation, which is also coherent with the hypothesis of the histone code.
The absence of a noticeable destabilization of the ORF8/DNA aggregate upon the inclusion of PTMs is also confirmed by the values of the binding free energy.
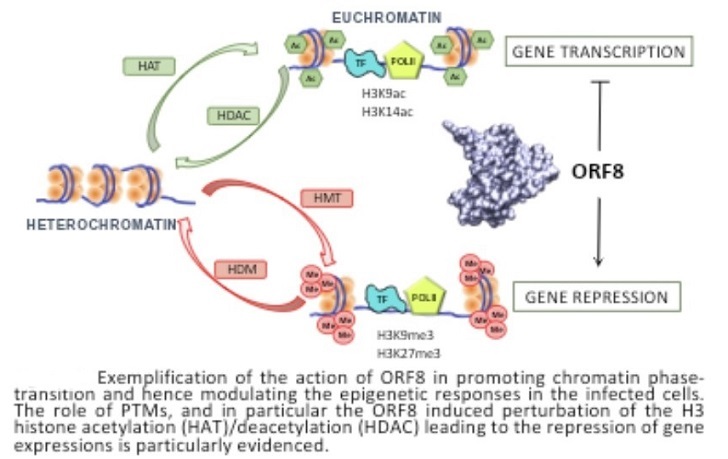
The study findings confirm the experimental evidences which reports that cells infected with viruses expressing ORF8 present a global more compact chromatin and, thus, an important under expression of various genes, especially those involved in the immune response.
While the findings are coherent with the role of histone mimic of ORF8, they can also help interpreting the interplay with PTMs.
The study findings have shown that ORF8 ARKS motif is able to maintain persistent interactions with DNA even in presence of methylation and acetylation.
It can therefore be surmised that, while ORF8 may effectively competes with the native H3 as a substrate for PTMs inducing enzymes, the introduction of acetylation and methylation on the viral protein will induce a different biological effect. Indeed, the presence of PTMs on ORF8 will not result in chromatin decompaction, hence it will perturb the epigenetic profile of the infected cell and thus favoring the viral reproduction. However, to confirm this hypothesis the interaction between ORF8 and acetylation and methylation enzymes should be properly modelled and simulated.
The study represents the first modelling of the epigenetic effects induced by a viral protein on the host cells. It helps in unravelling the complexes molecular mechanisms and cross-talks put in action by viruses to hijack the infected cell machinery and optimize their survival and reproduction.
For the latest
COVID-19 News, keep on logging to Thailand Medical News.

