BREAKING COVID-19 News! Iron-Sulfur Cluster Proteins Found In Viruses Including SARS-CoV-2 That Possibly Contributes To Pathogenesis!
Nikhil Prasad Fact checked by:Thailand Medical News Team Jan 19, 2024 1 year, 3 months, 1 week, 12 hours, 5 minutes ago
COVID-19 News: In the relentless pursuit of understanding the complex dynamics between viruses and host cells, recent breakthroughs in viral research have illuminated the involvement of iron–sulfur cluster proteins in viral pathogenesis. The Eunice Kennedy Shriver National Institute of Child Health and Human Development in Bethesda, USA, and Johns Hopkins University in the USA have jointly explored this fascinating aspect of virology. This
COVID-19 News report seeks to delve into the intricacies of iron–sulfur cluster proteins in viral genomes, with a special focus on the SARS-CoV-2 virus, shedding light on their potential contribution to the viral replication process.
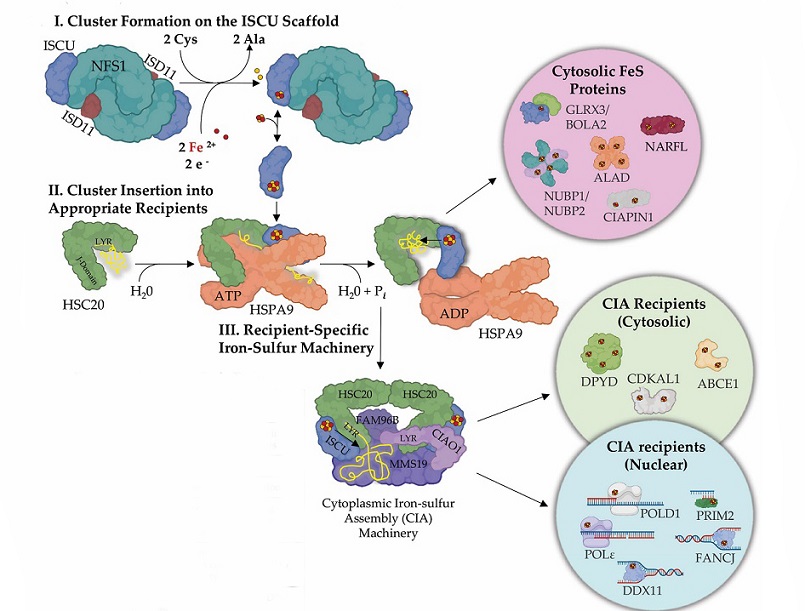 Iron–sulfur clusters and their assembly. Iron sulfur cluster biogenesis is a complex highly regulated process that requires two basic steps: (I) de novo FeS cluster formation on the main scaffold ISCU by the activity of a cysteine desulfurase, NFS1, which converts cysteine to alanine and generates a persulfide intermediate on ISCU in the process. Donation of iron and reducing equivalents completes [2Fe-2S] cluster assembly on the main scaffold ISCU. (II) The newly assembled FeS cluster is delivered to the appropriate recipient proteins by the activity of an HSP70 chaperone and cognate J-domain HSP40 cochaperone system. In mammalian cells, the chaperone is HSPA9 and the cochaperone HSC20 (aka HSCB). HSC20 recognizes LYR-like motifs present in subsets of FeS recipient proteins. In the cytosol of mammalian cells, following cluster incorporation, a subset of proteins, such as NARFL and CIAPIN1, complete their maturation and become active. (III) Nucleic acid processing enzymes require a highly specialized cytoplasmic iron–sulfur assembly (CIA) complex to acquire their cofactors. The CIA complex consists of CIAO1, MMS19, and FAM96B. CIAO1 was found to harbor a highly conserved LYR-motif that engages the cochaperone HSC20, thereby acting as a bridge between CIA and the de novo FeS assembly machinery. A subset of FeS proteins that acquire their cofactors from the CIA complex, (e.g., DPYD and ABCE1) may function in the cytosol, while others are translocated to the nucleus (POLD1, PRIM2, FANCJ).
The Significance of Iron–Sulfur Clusters in Cellular Functions
Iron–sulfur clusters and their assembly. Iron sulfur cluster biogenesis is a complex highly regulated process that requires two basic steps: (I) de novo FeS cluster formation on the main scaffold ISCU by the activity of a cysteine desulfurase, NFS1, which converts cysteine to alanine and generates a persulfide intermediate on ISCU in the process. Donation of iron and reducing equivalents completes [2Fe-2S] cluster assembly on the main scaffold ISCU. (II) The newly assembled FeS cluster is delivered to the appropriate recipient proteins by the activity of an HSP70 chaperone and cognate J-domain HSP40 cochaperone system. In mammalian cells, the chaperone is HSPA9 and the cochaperone HSC20 (aka HSCB). HSC20 recognizes LYR-like motifs present in subsets of FeS recipient proteins. In the cytosol of mammalian cells, following cluster incorporation, a subset of proteins, such as NARFL and CIAPIN1, complete their maturation and become active. (III) Nucleic acid processing enzymes require a highly specialized cytoplasmic iron–sulfur assembly (CIA) complex to acquire their cofactors. The CIA complex consists of CIAO1, MMS19, and FAM96B. CIAO1 was found to harbor a highly conserved LYR-motif that engages the cochaperone HSC20, thereby acting as a bridge between CIA and the de novo FeS assembly machinery. A subset of FeS proteins that acquire their cofactors from the CIA complex, (e.g., DPYD and ABCE1) may function in the cytosol, while others are translocated to the nucleus (POLD1, PRIM2, FANCJ).
The Significance of Iron–Sulfur Clusters in Cellular Functions
Iron-sulfur (FeS) clusters, essential cofactors in cellular processes, play a pivotal role in a myriad of biological functions across diverse organisms, from bacteria to humans. These clusters are involved in crucial activities such as mitochondrial respiration, the tricarboxylic acid (TCA) cycle, nitrogen fixation, DNA replication and repair, protein translation, heme and cofactor biosynthesis, and the regulation of iron levels. Their structural and functional versatility, enabling electron transfer processes, oxidoreductive reactions, and protein conformational changes, distinguishes FeS clusters from other metal ions.
Despite their ubiquity, the study of FeS clusters is challenging due to their susceptibility to oxidative destabilization in oxygen-rich environments. Mammalian cells, for example, typically maintain low oxygen levels, complicating the handling and study of FeS proteins. Advanced techniques such as
UV-vis absorption, Mössbauer and electron paramagnetic resonance (EPR) spectroscopies, nuclear magnetic resonance (NMR), electron-nuclear double resonance (ENDOR), Raman spectroscopy, native mass spectrometry (MS), and time-resolved native MS are employed to unravel the complexities of FeS clusters.
FeS clusters are not only structurally conserved throughout evolution but are also involved in vital cellular processes. Key cellular components such as the bacterial NADH dehydrogenase complex and eukaryotic mitochondrial respiratory complex I contain conserved FeS clusters that contribute to electron transfer and proton pumping. Additionally, DNA replication proteins, including DNA polymerases and repair enzymes, rely on FeS clusters for genome maintenance and cell proliferation.
Biogenesis of FeS Clusters and their Transfer to Proteins
Cells have evolved intricate biosynthetic pathways for the assembly of FeS clusters and their incorporation into various proteins. In mammalian cells, the rhombic [2Fe-2S] cluster, the basic building block of all FeS clusters, is assembled on the main scaffold protein, ISCU. This assembly involves the cysteine desulfurase NFS1, which converts cysteine to alanine while mobilizing inorganic sulfur for FeS cluster assembly. The iron donor for FeS cluster assembly in mitochondria remains a subject of debate, with PCBP1 and its binding partner BOLA2 identified as contributors in the cytosol.
Transfer of newly assembled FeS clusters to recipient proteins is facilitated by an evolutionarily conserved chaperone/co-chaperone system. In humans, the ATP-dependent Hsp70 chaperone, HSPA9, and its cognate J-domain protein, HSC20, play a crucial role in this process. The co-chaperone HSC20 recognizes specific motifs in recipient FeS apoproteins, ensuring the specificity of FeS cluster allocation. The Cytoplasmic Iron–sulfur Assembly (CIA) complex is also involved in the acquisition of FeS clusters by a subset of nucleocytoplasmic FeS proteins.
FeS Biogenesis Pathway and Human Health
Recent years have witnessed the identification of ultra-rare disorders caused by loss-of-function mutations in genes encoding essential FeS biogenesis proteins. Disorders associated with deficiencies in components of the CIA machinery, such as CIAO1, highlight the critical role of FeS clusters in genome stability, DNA metabolism, and various physiological systems. The exploration of these pathways through advanced genetic techniques like exome sequencing has contributed to our understanding of rare human conditions and potential therapeutic targets.
Iron–Sulfur Clusters in Viruses
Unlike bacterial pathogens, many viruses have evolved as genetic minimalists, relying on host cell machinery for genome replication and transcription. FeS clusters, crucial for numerous cellular functions, have been identified in viral proteins, suggesting their involvement in viral replication processes. This study focused on five diverse virus families, including the SARS-CoV-2 virus, known to encode FeS proteins.
SARS-CoV-2 and FeS Proteins: Unraveling the Replication Complex
The virus responsible for the global COVID-19 pandemic, SARS-CoV-2, belongs to the Nidovirales order and is distinguished by its large single-stranded RNA genome. Notably, SARS-CoV-2 encodes two FeS proteins, nsp12 and nsp13, crucial components of the replication and transcription complex (RTC). The discovery of FeS clusters in these viral proteins garnered attention during the pandemic, as it revealed a previously overlooked aspect of the virus's replication machinery.
Nsp12, the RNA-dependent RNA polymerase (RdRp), is the catalytic subunit of the RTC and contains two [4Fe-4S] clusters in its structure. These clusters, initially mischaracterized as zinc centers, were found to be essential for replication, with one cluster at the interface between the NiRAN and catalytic domains and the second in the fingers subdomain within the polymerase domain. The FeS clusters enhance the polymerization efficiency and binding affinity to RNA, contributing significantly to the replication process.
Nsp13, the helicase enzyme of SARS-CoV-2, also ligation [4Fe-4S] clusters and Zn2+ ions. The FeS cluster in nsp13 enhances its binding selectivity for RNA over DNA, a crucial factor in its unwinding activity. The coexistence of diverse metal compositions in nsp13 is a unique feature and underscores the complexity of FeS utilization in viral proteins.
Targeting FeS Clusters for Antiviral Strategies
Exploiting the vulnerability of FeS clusters to oxidative damage, studies have explored the potential of using FeS-targeting agents as antiviral therapeutics. TEMPOL, a stable nitroxide small molecule, has demonstrated efficacy in inhibiting SARS-CoV-2 replication by inducing oxidative degradation of FeS clusters in viral enzymes. This innovative approach offers a promising avenue for developing antiviral drugs that selectively target viral replication machinery while sparing host cell functions.
Conclusions and Future Perspectives
The exploration of iron-sulfur cluster proteins in viral genomes, particularly in SARS-CoV-2, has unveiled a new dimension in our understanding of viral replication mechanisms. The evolutionary conservation of FeS-ligating centers in viral proteins underscores their importance in enhancing viral fitness. Moreover, the vulnerability of FeS clusters to oxidative damage presents an exploitable target for antiviral interventions.
As we stand at the forefront of an exponential discovery phase, the identification of novel FeS proteins in viruses may open doors to innovative antiviral strategies. Further research into the conservation and evolution of viral FeS proteins across different virus families and host organisms will deepen our understanding of the role these clusters play in viral infections. The intersection of virology, bioinorganic chemistry, and drug discovery holds immense potential for shaping the future landscape of antiviral therapeutics and advancing our knowledge of fundamental aspects of viral biology.
The study findings were published in the peer reviewed journal: Inorganics.
https://www.mdpi.com/2304-6740/12/1/34
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
