BREAKING NEWS! COVID-19 Genomics: Emory University Research Warns Emerging Recombinant SARS-CoV-2 Strains Already Occurring
Source: COVID-19 Genomics Aug 07, 2020 5 years, 4 months, 2 weeks, 1 day, 16 hours, 49 minutes ago
COVID-19 Genomics: Researchers from Emory University-Atlanta have confirmed that recombination events among the viral strains of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are already occurring and have detected the presence of such recombinant strains.
.jpg)
The research findings were published on a preprint server and are currently under peer-review.
https://www.biorxiv.org/content/10.1101/2020.08.05.238386v1
The causative virus SARS-CoV-2 has been under intense research and epidemiological scrutin primarily due to the potential of viral recombination, which can result in viral genotypes with modified phenotypic characteristics – including altered transmissibility, pathogenicity, and virulence.
The propensity for recombination among the wider group of Betacoronaviruses is well established hence the need to be on a constant lookout.
A rather small number of polymorphic, phylogenetically informative sites in the SARS-CoV-2 genome implies that detecting recombinant genomes is cumbersome and highly dependent on the identity of the parent clades.
But even despite these drawbacks, two studies have already reported recombinants among different SARS-CoV-2 strains.
Researchers from Emory University and Emory-UGA Center of Excellence for Influenza Research and Surveillance (CEIRS) in Atlanta (United States) decided to revisit these findings by utilizing a new analysis approach in order to elucidate potential recombination within SARS-CoV-2.
The study employed a four-step approach to pinpoint recombinant SARS-CoV-2 genomes. First, mutations that define the clonal pattern of inheritance were characterized, followed by the identification of genomes that violate this pattern. Then the researchers identified and refined the boundaries of genetic transfer and finally assessed the plausibility of transfer by determining potential co-circulation of predicted parental clades.
Please Help To Donate To Sustain This Site And Other Research We Are Propelling. Thank You. https://www.thailandmedical.news/p/sponsorship
Various genomes were downloaded from the GISAID genome databases and subsequently filtered to exclude low-quality sequences. Furthermore, genomes were aligned in accordance with the NCBI reference sequence genome using a multiple sequence alignment program known as MAFFT.
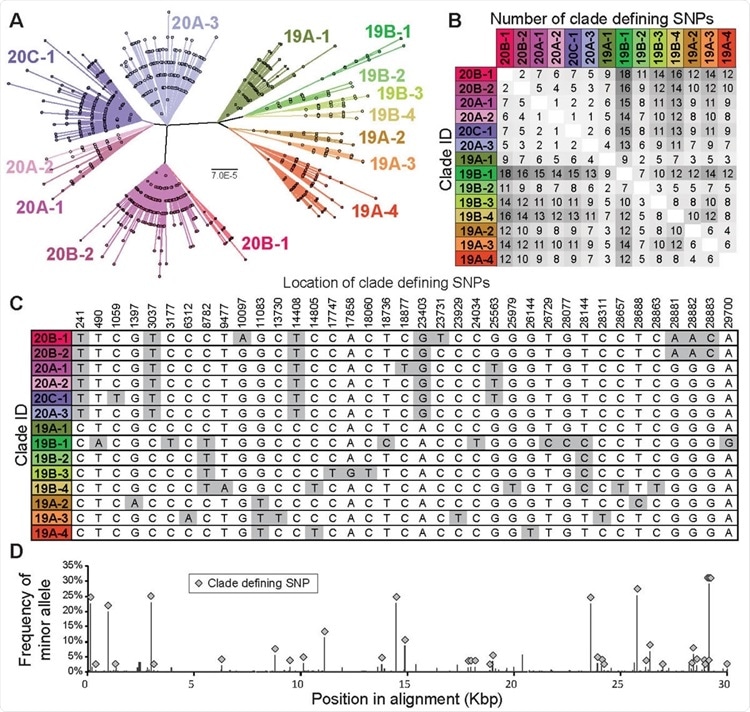 The clade structure of SARS-CoV-2 is structured predominantly by 37 clade-defining SNPs. (A) Maximum likelihood phylogeny based on the General Time Reversible model with invariant sites of 9783 high quality unique genome sequences with <1% Ns. 14 monophyletic clades were identified manually. These clades gener
ally correspond to SARS-CoV-2 clades defined in Nextstrain (Hadfield et al., 2018), although a fraction of them are at higher resolution than Nextstrain clades. Clades defined here are named by Nextstrain clade designation (e.g., 20B) followed by a subclade number (e.g., -1). Scale bar is in substitutions per site. (B) Pairwise differences between the clade-defining SNP profiles of all 14 clades. (C) Location and nucleotide identity of clade-defining SNPs and (D) their frequency among SARS-CoV-2 genomes.
The clade structure of SARS-CoV-2 is structured predominantly by 37 clade-defining SNPs. (A) Maximum likelihood phylogeny based on the General Time Reversible model with invariant sites of 9783 high quality unique genome sequences with <1% Ns. 14 monophyletic clades were identified manually. These clades gener
ally correspond to SARS-CoV-2 clades defined in Nextstrain (Hadfield et al., 2018), although a fraction of them are at higher resolution than Nextstrain clades. Clades defined here are named by Nextstrain clade designation (e.g., 20B) followed by a subclade number (e.g., -1). Scale bar is in substitutions per site. (B) Pairwise differences between the clade-defining SNP profiles of all 14 clades. (C) Location and nucleotide identity of clade-defining SNPs and (D) their frequency among SARS-CoV-2 genomes.
Also clades were identified as monophyletic groups situated within a maximum likelihood phylogenetic tree built from 9,783 unique high quality genome sequences by employing PhyML (i.e., a software package that analyzes alignments of nucleotide or amino acid sequences in a phylogenetic framework with the use of modern statistical approaches).
To visualize the empirical support for recombination, the researchers performed phylogenetic analysis on subsets of the SARS-CoV-2 viral genome, which correlate to stretches of the genome bounded by inferred regions of transfer.
They aimed to appraise, based on geographic considerations, the feasibility of required transfer events between the predicted parental clades to generate the recombinants that were observed in this study. The results they have obtained were rather insightful.
The study revealed recombination in five SARS-CoV-2 genomes
Dr Anice C. Lowen, Professor at the Department of Microbiology and Immunology, Emory University, Atlanta told Thailand Medical News, "In total, we screened 47,390 unique genomes and identified five genomes that are strong candidates for having evolved through recombination between two distantly related parental clades.”
The sequences were linked to infections from the United States, United Kingdom and from China; furthermore, each of these genomes harbors phylogenetic markers of two distinct SARS-CoV-2 clades, and each recombinant genome was found to cluster tightly with the predicted parent clades across the regions of transfer.
Taking into consideration the latter, the predicted parent clades of aforementioned recombinant genomes were (with one exception) reported to be co-circulating in the country of infection in the two weeks before to the sample collection process.
Dr Lowen, a co-researcher of the study added, "By identifying the nucleotide changes that underpin the clonal phylogeny of SARS-CoV-2, we established criteria for identifying putative recombinant genomes, and for evaluating their plausibility.”
The researchers stressed the need to monitor high-fitness recombinants.
The study team commented, "Ultimately, our results suggest that recombination between SARS-CoV-2 strains is occurring, but these chimeric genotypes remain rare. As the pandemic continues to expand, the population genetic diversity of SARS-CoV-2 will increase, making it easier to detect recombinant genomes.”
The new recombinant genomes the researchers have identified in this study may not belong to persistent, larger recombinant lineages. Instead, they may represent fleeting observations into lineages that were not successfully established or have gone extinct.
However the growing number of mutations will also increase the possibility that recombinant genomes exhibit modified phenotypic characteristics that can impact fitness. On the other hand, transmission heterogeneity means there is a much lower chance for any given viral infection to form a persisting lineage.
The key thing however is that this study shows that recombination is already taking place in SARS-CoV-2, real-time analyzes and increased surveillance endeavors (such as this one) should be encouraged in order to monitor the circulation and possible spread of high-fitness recombinant viral genotypes, the researchers warn.
For more on
COVID-19 Genomics, keep on logging to Thailand Medical News.
Please Help To Donate To Sustain This Site And Other Research We Are Propelling. Thank You. https://www.thailandmedical.news/p/sponsorship
.jpg)
