BREAKING! Ohio State University Discovers That SARS-CoV-2 BQ.1 And BQ.1.1 Variants Exhibit Enhanced Fusogenicity Besides Increased Immune Evasiveness!
Source: COVID-19 News -BQ.1, BQ.1.1 - Increased Fusogenicity Oct 21, 2022 2 years, 5 months, 4 weeks, 2 days, 1 hour, 7 minutes ago
COVID-19 News: In a new research by scientists from The Ohio State University – USA and Wexner Medical Center, Ohio - USA to study the neutralization resistance of the subvariants a BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2 as well as their ancestral BA.4/5, BA.2.75 and D614G variants against sera from 3-dose vaccinated health care workers, hospitalized BA.1-wave patients, and BA.5-wave patients, the study team unexpectedly discovered that the BQ.1 and BQ.1.1 variants exhibit enhanced fusogenicity besides increased immune evasiveness.
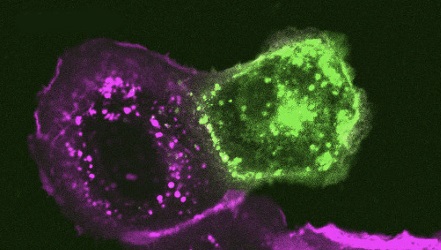
In order to examine how alterations to the S protein in these new Omicron subvariants impact function, the study team examined the ability of the S from these subvariants to mediate cell-cell fusion. Consistent with prior Omicron subvariants, all new subvariants exhibited diminished syncytia formation compared to ancestral D614G, despite comparable levels of cell surface expression of each S construct. Notably, the new Omicron subvariants BA.4.6, BQ.1, and BQ.1.1 exhibited enhanced fusogenicity compared to their parental variant BA.4/5, and similarly, BA.2.75.2 showed an increased fusogenicity relative to BA.2.75.
Considering the key role of cellular furin in processing viral S protein into S1 and S2 subunits for SARS-CoV-2 entry, the study team next examined the proteolytic processing of S in lentivirus producer cells.
They saw an enhanced S2/S ratio compared to ancestral D614G in BA.4.6, BF.7, BQ.1, and BQ.1.1 with a 1.8-fold, 2.2-fold, 3.6-fold, and 2.8-fold higher S2/S ratio, respectively, compared to the 1.6-fold higher S2/S ratio in BA.4/5.
However, compared to BA.4/5, the S1/S ratio of these BA.4/5-derived subvariants was comparable (BF.7 and BQ.1.1) or even reduced (BA.4.6 and BQ.1), suggesting possible shedding of S1 into the culture media. BA.2.75 and BA.2.75.2 exhibited minimal difference in S2/S ratio compared to D614G, but once again, BA.2.75.2 exhibited a decreased S1/S 216 ratio compared to the parental BA.2.75, suggesting possibly increased S1 shedding for the BA.2.75.2 subvariant.
The study findings were published on a preprint server ad are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2022.10.19.512891v1
The study findings also showed enhanced neutralization resistance in all new subvariants, especially the BQ.1 and BQ.1.1 subvariants driven by a key N460K mutation and to a lesser extent, R346T and K444T mutations, as well as the BA.2.75.2 subvariant driven largely by its key F486S mutation.
The BQ.1 and BQ.1.1 subvariants exhibited enhanced fusogenicity and S processing dictated by the key N460K mutation.
Interestingly, the BA.2.75.2 subvariant saw an enhancement by the F486S mutation and a reduction by the D1199N mutation to its fusogenicity and S processing resulting in minimal overall change.
Molecular modelling revealed the mechanisms of receptor-binding and non-receptor binding monoclonal antibody-mediated immune evasion by R346T, K444T, F486S and D1199N mutations.
The study findings also showed that The N460K and F486 mutations enhance fusogenicity while D1199N diminishes fusogenicity.
The study team told
ws/">COVID-19 News reporters at Thailand
Medical News,” In particular, the strong evasion of BA.4/5 infection-induced sera is concerning, as the recently recommended bivalent mRNA vaccine boosters contain BA.4/5 S along with the prototype. This possibility, together with the emergence of more diverse SARS-CoV-2 variants necessitates the urgent development of more broadly active, even pan-coronavirus, COVID-19 vaccines.”
The study team added, “As circulating SARS-CoV-2 diversifies, the ability to vaccinate against dominant circulating variants may be even more compromised. Hence, examination of the efficacy of bivalent mRNA vaccines against emerging variants is critical to control and end the global pandemic.”
The team added, “These new Omicron subvariants also affect the fusogenicity of the S proteins. Similar to previous finding that BA.4/5 and BA.2.75 exhibit increased fusion capability, here we also observed increased cell-cell fusion in several new Omicron subvariants compared to their respective parental subvariants, BA.4/5 or BA.2.75. These data may together indicate a continuing shift towards more efficient transmembrane protease, serine 2 (TMPRSS2) utilization to allow for plasma membrane mediated viral fusion and entry pathway. Consistent with this notion, we observed an increased furin-cleavage efficiency in several S proteins, especially BQ.1, and BQ.1.1 subvariants, which could improve TMPRSS2 utilization.”
The researchers warned that this is concerning, because previously it has been shown that the shift in entry pathway from TMPRSS2-mediated plasma membrane fusion used by the prototype SARS-CoV-2 towards cathepsin B/L-mediated endosomal entry observed in the Omicron clade is associated with reduced lung tropism, increased nasopharyngeal tropism, and reduced pathogenicity of the Omicron variant.
That said, the study findings show that the new Omicron subvariants continue to exhibit weak infection efficiency in lung-derived CaLu-3 cells. Examination of tissue tropism and pathogenicity in vivo for these and future emerging Omicron subvariants is critical to tailor appropriate public health responses.
The study team concluded, “The perpetual emergence of SARS-CoV-2 variants with enhanced immune escape continues to threaten public health. Monitoring the immune escape of emerging variants will be critical to improving mRNA vaccine reformulation, assessing new broader coronavirus vaccine candidates, as well as directing ongoing public health measures. Further, emerging variants must be monitored closely for any indication of selective pressure in enhancing lung tropism and potentially pathogenicity to ensure that any highly transmissible and more pathogenic variants are better and more quickly contained. Altogether, these findings shed light on the concerning evolution of newly emerging SARS-CoV-2 Omicron subvariants.”
It should also be noted that the BQ.1 and BQ.1.1 sub-lineages are still aggressively evolving and spawning more sub-variants, At present a total of 36 sub-lineages have been identified including BQ.1.1.1 to BQ.1.1.10, BQ.1.2 to BA.1.20, BQ.2, BQ.1.8.1, BQ.1.8.2, BQ.1.10.1 etc.
https://cov-spectrum.org/collections/24
.jpg)
For the latest
COVID-19 News, keep on logging to Thailand
Medical News.

.jpg)