BREAKING! Prospective ‘Omega’ SARS-CoV-2 Variant Has Reemerged In Time For Fall And Winter! The R.1 Variant From Japan, Now In U.S. And Spreading Globally!
Source: SARS-CoV-2 R.1 Variant Sep 23, 2021 3 years, 7 months, 1 week, 3 hours, 9 minutes ago
R.1 Variant: A new SARS-CoV-2 variant called R.1 has reemerged after months of gaining viral fitness and relevant mutations on its genome to make it truly virulent, potent, extremely transmissible and immune evasive. The R.1 may be easier to contract due to protein mutations within the spike protein of the virus and its ability to be easily transmitted from person to person despite vaccination status does have scientists on edge.
Although this variant was first identified in Tokyo, Japan early as December 2020,
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009619
https://onlinelibrary.wiley.com/doi/10.1002/jmv.27240
https://www.medrxiv.org/content/10.1101/2021.03.16.21253248v1
it had not gained much mutations in its genome yet and coupled the fact that the arrogant Western world and Western researchers typically are not interested in what is developing in Asia, not many were paying careful attention to the developments of this new identified variant. It also did not help that the GISAID and Nextstrain platforms have limitations in reporting and accurately detecting mutation developments on emerging SARS-CoV-2 variants in great detail.
The variant was also detected by French researchers in Marseille between February and April 2021.
https://www.mediterranee-infection.com/wp-content/uploads/2020/04/Ms_Colson_ArchVirol_Jul2021_vL_IHUPreprint.pdf
A former Harvard Medical School professor and infectious disease expert Dr William A Haseltine was the only western scientists paying careful attention to this variant and studying in detail its evolution and mutation changes along with experts from the Access Health International platform.
So now in late September 2021, at the start of fall, we finally have the emergence of variant that has gained the necessary viral fitness and is fast spreading around the whole with very little attention being paid to it and the inadequate genomic surveillance by many countries is also helping in its spread.
The R.1 variant was easily replaced by the Alpha and Delta variants but has now because of evolution and gained viral fitness , been revived and is expected to replace the Delta variant in a matter of weeks.
From the analysis of the GISAID SARS-CoV-2 database, the variant has infected over 10,000 people globally and is fast spreading and has already been detected in various States in the U.S. including in a nursing home in Kentucky where it infected 45 residents and healthcare workers.
The new variant is related to the original coronavirus strain and contains "many unique mutations" that can lead to “increased resistance to antibodies in convalescent sera and to neutralizing antibodies that empowers it to bypass antibodies in a victim.
It h
as been found that the evolved variant was successful in affecting multiple vaccinated people by circumventing antibodies.
The R.1.variant shares the highly infectious D614G mutation that is present in other variants of concern such as the Delta, Alpha, Beta and Gamma.
The R.1.variant contains five mutations previously noted in variants of concern or interest, two of which are in the Spike protein. It also contains many unique mutations.
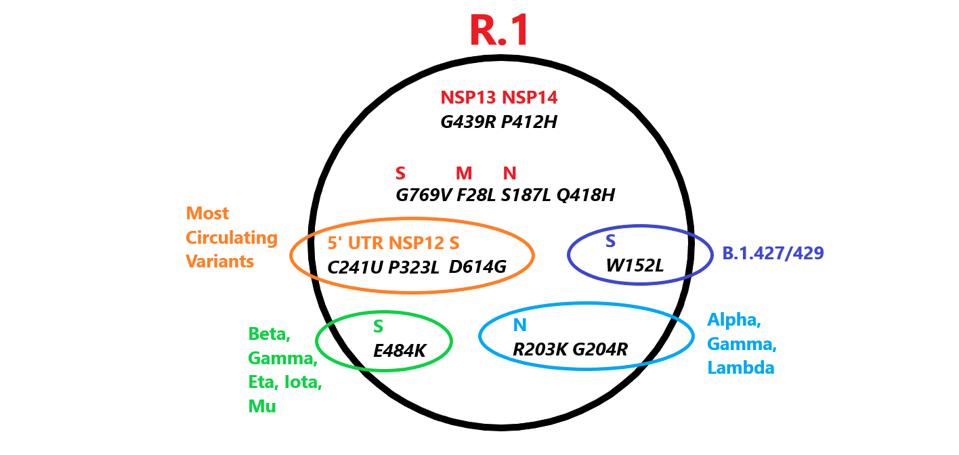 Mutations in R.1 found in other variants. Those in red are unique to R.1. Credit: Access Health International
Mutations in R.1 found in other variants. Those in red are unique to R.1. Credit: Access Health International
The R.1 variant also shares a common origin with all variants of interest or concern. Three mutations are common in all these VOIs, VOCs and the R.1 variant ie:
1. The mutation C241U found in the 5’ untranslated region: C241U,
2. The mutation P323L found in the viral polymerase NSP12
3. The mutation D614G found in the exterior S1 domain of the spike protein.
It has been found that the D614G mutation increases viral infectivity but the significant of the other two are not known. Together these three mutations characterize the first major variant first observed in early 2020. However that that virus soon displaced almost all of the original Wuhan isolates. The ‘Triad’ virus is the parent of all variants of concern or, interest including Alpha, Beta, Gamma, Delta, Lambda, and Mu. Only two currently circulating variants, A.30 and A.23.1, both of East Africa origin, lack the Triad and are most likely independent descendants of the original Wuhan virus.
Spike Mutations Found In R.1. That are Immune Evasive
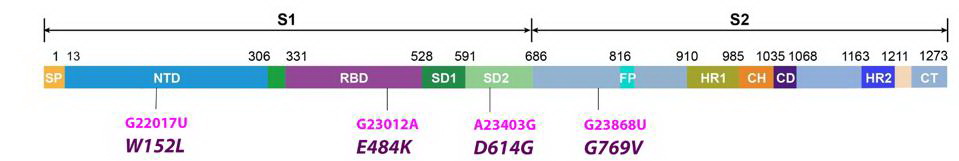
It is already known that the spike protein of SARS-CoV-2 binds the host ACE2 receptor and initiates infection. The R.1 variant contains three Spike mutations, two of which are previously noted in other variants the other which is unique.
The R.1 variant has the E484K mutation in the frequently mutated domains of the spike protein ie the receptor-binding domain (RBD).
 Common mutations in the nonstructural proteins in the R.1 variant genome. Color code: nonsynonymous mutations (pink) and their corresponding amino acid mutations (purple) Credit: Access Health International
Common mutations in the nonstructural proteins in the R.1 variant genome. Color code: nonsynonymous mutations (pink) and their corresponding amino acid mutations (purple) Credit: Access Health International
Studies have shown that the 484 mutation conveys increased resistance to antibodies in convalescent sera and to neutralizing monoclonal antibodies. The E484K substitutes the negatively charged residue glutamic acid for the positively charged lysine. The E484K is present in the Beta, Gamma, Eta, Iota, and Mu variants.
The R.1 variant also contains the W152L mutation in the N-terminal domain. This region of the spike protein is also the target of neutralizing antibodies in convalescent sera and is the target of several neutralizing monoclonal antibodies. An identical mutation at position 152 is present in one of the sub-variants of the Delta strain found in India. The very same amino acid is mutant in several variants first detected in California, B.1427/429. The 152 tryptophan is substituted for leucine in the California variants. Multiple independent mutations of the same amino acid imply functional advantage.
The newly discovered mutation, G769V, falls in an interdomain region between positions 732 and 780. It is unique to the R.1 variant and its functional characteristics are not yet known.
The Non-Structural Proteins Of R.1
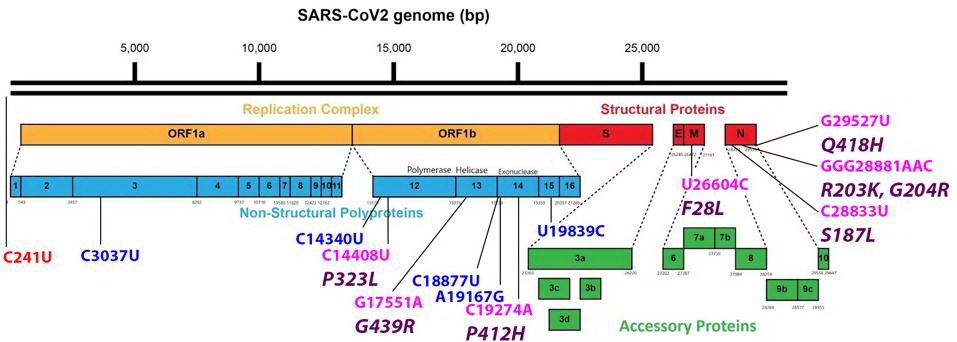 Common mutations external to the S protein in the R.1 variant genome. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations (purple). Credit: Access Health International
Common mutations external to the S protein in the R.1 variant genome. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations (purple). Credit: Access Health International
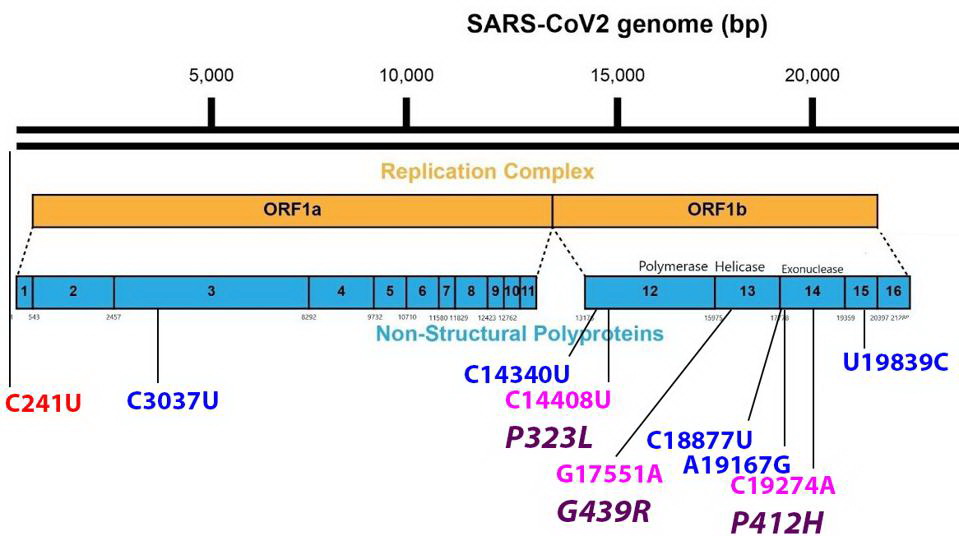 Mutations found in the Orf1ab non-structural proteins. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations (purple). Credit: Access Health International
The G439R Mutation On The R.1 Non-Structural Protein 13 (NSP13)
Mutations found in the Orf1ab non-structural proteins. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations (purple). Credit: Access Health International
The G439R Mutation On The R.1 Non-Structural Protein 13 (NSP13)
The
Non-Structural Protein 13 or NSP13 protein is the helicase and part of the replication transcription complex. The G439R mutation of R.1 is unique to this variant. The 439 residue lies adjacent to an active ATP-binding site. The Glycine to arginine mutation is a change from a neutral amino acid to one that carries a positive charge. The effect of this mutation on replication and helical activity is of interest to scientists.
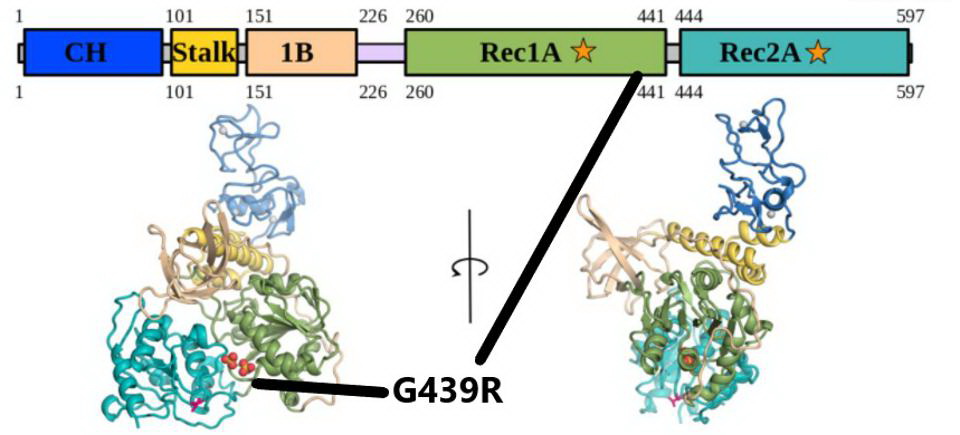 The G439R mutation in NSP13. The mutation falls in the Rec1A domain, noted by the black lines. Credit: White et al
The G439R mutation in NSP13. The mutation falls in the Rec1A domain, noted by the black lines. Credit: White et al
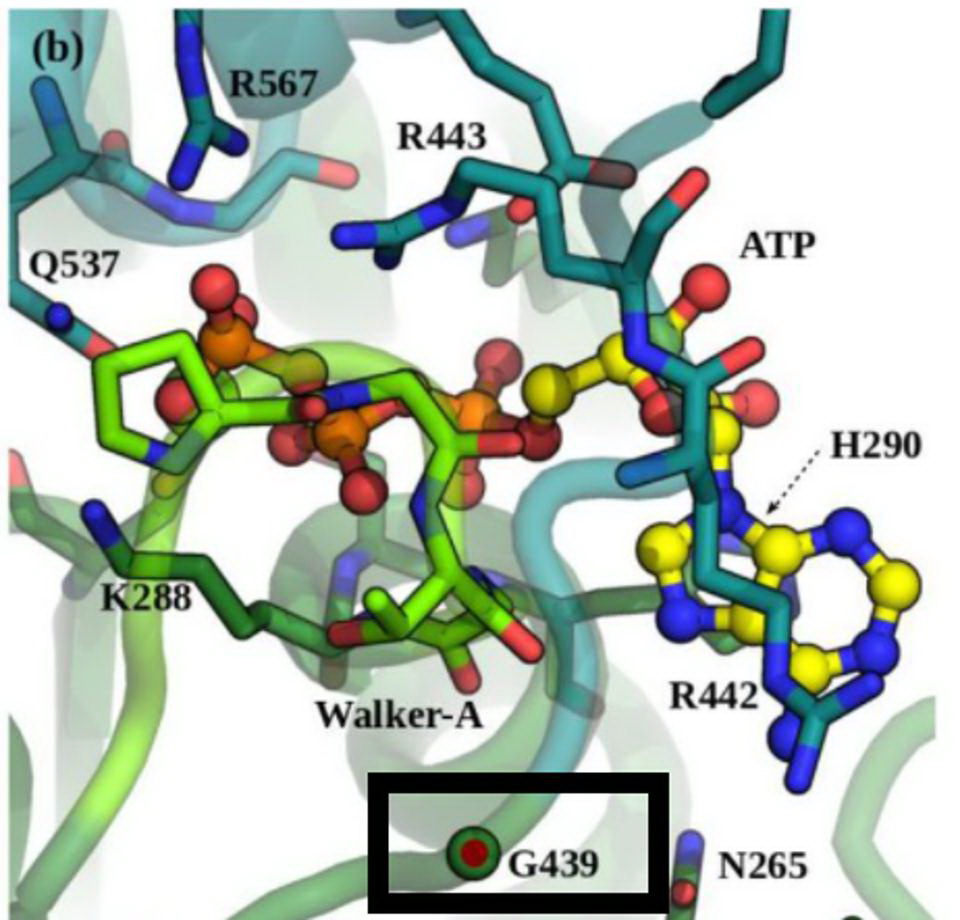 Position G439 located near the ATP-binding site in NSP13. Credit: White et al
The P412H mutation On The R.1 Non-Structural Protein 14 (NSP14)
Position G439 located near the ATP-binding site in NSP13. Credit: White et al
The P412H mutation On The R.1 Non-Structural Protein 14 (NSP14)
The NSP14 protein is an exoribonuclease and like the NSP13 helicase, is a part of the replication transcription complex. The exoribonuclease has an error-editing function and also confers resistance to chain-terminating nucleotides. The NSP14 protein of R.1 carries the unique P412H mutation. NSP14 is also is a methyltransferase that participates in the formation of capped viral messenger RNAs. The capping of viral messenger RNAs is required to circumvent triggering the innate immune response. A change from the amino acid proline to histidine substitutes a neutral amino acid for one that carries a positive charge, again indicating a major charge shift and may enhance the properties of the protein. The site of the mutation lies within the N7 methyltransferase capping domain of the protein adjacent to SAH and GpppA binding.
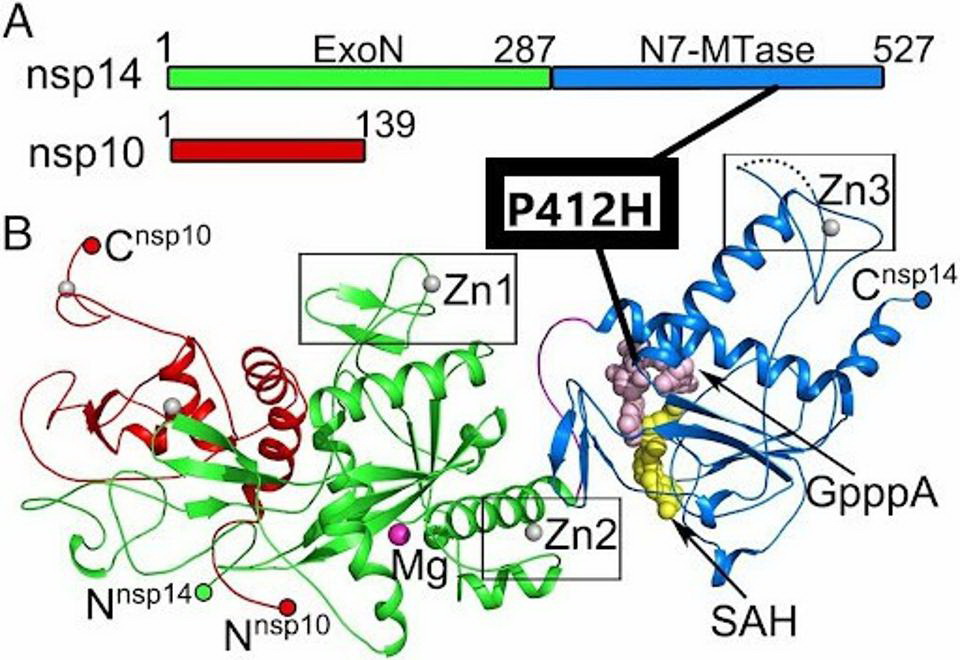 The P412H mutation in NSP14. The mutation is located in the N7-MTase subdomain near SAH and GpppA binding sites. Credit: Yuanyuan et al
Unique Synonymous Nucleotide Changes Found In R.1
The P412H mutation in NSP14. The mutation is located in the N7-MTase subdomain near SAH and GpppA binding sites. Credit: Yuanyuan et al
Unique Synonymous Nucleotide Changes Found In R.1
It has also been observed there are a number of synonymous nucleotide changes, which could impact the RNA replication and transcription processes. In the nonstructural proteins, it was found the following nucleotide changes C3037U, C14340U, C18877U, A19167G, and T19839C. It is possible that, while these do not change amino acids, they could change the secondary structure of the RNA which may result in functional consequences.
The R.1.Structural Proteins
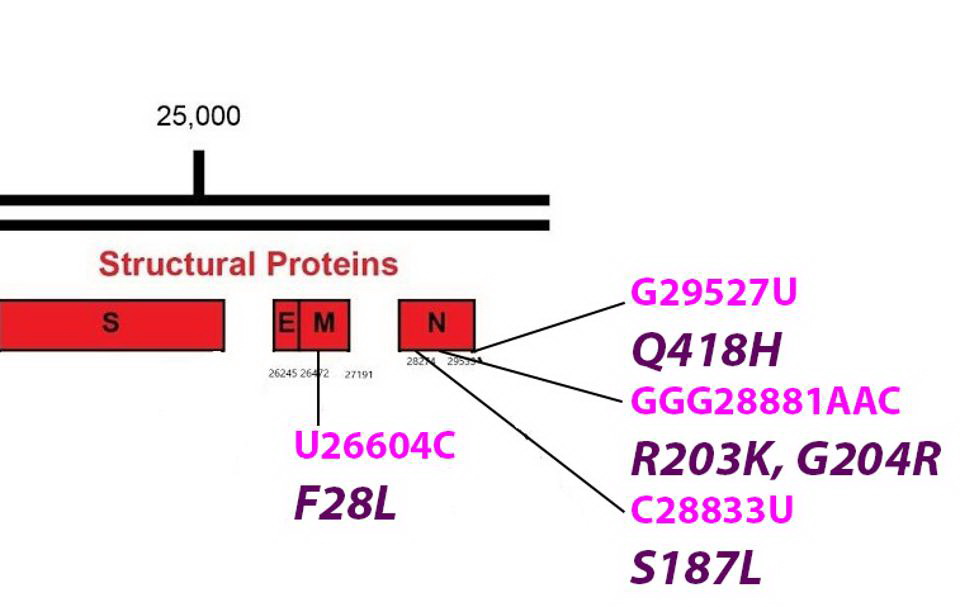 Common mutations in the non-Spike structural proteins in the R.1 variant genome. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations. Credit: Access Health International
The F28L Mutation Found In The R.1 The Membrane Protein (M)
Common mutations in the non-Spike structural proteins in the R.1 variant genome. Color code: noncoding nucleotide mutations (red), synonymous mutations (blue), nonsynonymous mutations (pink), and their corresponding amino acid mutations. Credit: Access Health International
The F28L Mutation Found In The R.1 The Membrane Protein (M)
The viral membrane protein is key to the formation of virus particles. It is embedded in the membrane that surrounds the virus. M is reported to assist in the packaging of viral into mature virions. M also is active in the suppression of innate immunity. The F28L is unique to R.1. The amino acid change lies within the first of three transmembrane regions domains. Phenylalanine and leucine are both nonpolar and uncharged amino acids. Nonetheless, the variant could impact the multiple functions of M including the stability of the virus particle.
.jpg) The F28L mutation in the M protein. The mutation is found in a transmembrane region (TMR).Credit: Access Health International
Some scientists speculate that all initial antiviral compounds that were found to target the membrane protein of the initial wildtype Wuhan strain could be rendered ineffective because of this new mutation on the membrane protein.
Various Mutations Found On the R.1 Nucleocapsid Protein (N)
The F28L mutation in the M protein. The mutation is found in a transmembrane region (TMR).Credit: Access Health International
Some scientists speculate that all initial antiviral compounds that were found to target the membrane protein of the initial wildtype Wuhan strain could be rendered ineffective because of this new mutation on the membrane protein.
Various Mutations Found On the R.1 Nucleocapsid Protein (N)
The
Nucleocapsid Protein or N is the most active of the virus structural proteins. N wraps the viral genome into a helical structure and is required for replication and transcription. During early infection, N is reported to play an important role in the suppression of innate immunity. N is highly antigenic and most infected individuals have significant titers of anti-N antibodies.
.jpg) R.1 mutations in the N protein, found in the linker and C-terminal domain, followed by a 3D representation. Credit: Cubuk et al
R.1 mutations in the N protein, found in the linker and C-terminal domain, followed by a 3D representation. Credit: Cubuk et al
The R.1 variant has mutations on the protein. R.1 shares the R203K and G204R mutations with three variants of concern Alpha, Gamma, and Lambda and with other reported variants as well. The amino acids change in the neighboring amino acid at positions 203 and 204 are always found as a pair. The two mutations lie near the center of the linker domain that connects the RNA binding domain of N with the dimerization domain. The change from glycine to arginine at position 204 introduces a second positive charge into a region that is predominantly neutral.
Another mutation found on the R.1. N protein is the S187L mutation located in the linker domain.
Yet another mutation found on the N protein of the R.1. ie the Q418H mutation adds a positive charge to the extreme carboxy terminus of N.
R.1 is a variant to watch. It has established a foothold in both Japan and the United States. In addition to several mutations notably in the spike and nucleocapsid protein in common with variants of concern, R.1 has a set of unique mutations that may confer an additional advantage in transmission, replication, and immune suppression.
Still Evolving
Scientist are still studying the R.1 variant has there are many more mutations found on that are not covered in this article yet.
In terms of evolution rate, compared to the Delta variant that is spawning out many new sub-variants, the R.1 is mutating even more faster and is striving for absolute viral fitness.
New infections with the reemerged R.1 variant show a greater degree of disease severity requiring hospitalization and also a high mortality rate.
Spreading Fast But Quietly
Data from the site Outbreak.info, a website that collects data on COVID variants, as of September 23, R.1 has now infected more than 10,567 people around the world and has been detected in 47 states.
https://outbreak.info/
Maryland was found to have the highest number of cases, with 399 cases being detected since it was first found in the country.
In total, 2,259 Americans have been found with the R.1 strain which was first detected in the country on March 15, 2020. The latest case to be detected in the U.S. was on August 6 and was found in 0.5 percent of cases.
Outbreak.info also reported the strain has been found in at least 31 countries worldwide, including China, India and many nations in Western Europe.
The R.1 variant never caught the attention of researchers or public health officials till the work of Professor Haseltine and Access health International was made public a few days ago.
Researchers and health authorities have to really wake up and maintain more stringent genomic surveillance. A lot of emerging variants and mutations of concern are escaping their notice. Vaccine and drug manufacturers also need to kept abreast of all these developments. Doctors and other healthcare staff also need to be aware of what variants they are dealing with as the pathogenesis of each variant is unique and clinical manifestations may differ and require a different treatment protocols.
For more on the
R.1 Variant, keep on logging to Thai









.jpg)
.jpg)