BREAKING! Scientists Discover Blocking Protein Interactions Between Human G3BP1/2 Proteins And SARS-CoV-2 ΦxFG Peptide Motif Inhibits Infection.
Source: Medical News - SARS-CoV-2 Research Nov 24, 2021 3 years, 10 months, 3 weeks, 16 hours, 35 minutes ago
Scientists from University of Copenhagen-Denmark, Uppsala University-Sweden, Umeå University-Sweden, Karolinska Institutet-Sweden and the Institute of Cancer Research-UK have in a new study discovered a novel method of inhibiting SARS-CoV-2 infection in human cells by blocking certain protein interactions, specifically between human G3BP1/2 proteins and SARS-CoV-2 ΦxFG Peptide Motif.
Viral proteins make extensive use of short peptide interaction motifs to hijack cellular host factors. However, most current large-scale methods do not identify this important class of protein-protein interactions. Uncovering peptide mediated interactions provides both a molecular understanding of viral interactions with their host and the foundation for developing novel antiviral reagents.
The study team describe a viral peptide discovery approach covering 23 coronavirus strains that provides high resolution information on direct virus-host interactions.
The study findings identified 269 peptide-based interactions for 18 coronaviruses including a specific interaction between the human G3BP1/2 proteins and an ΦxFG peptide motif in the SARS-CoV-2 nucleocapsid (N) protein.
Importantly this interaction was found to support viral replication and through its ΦxFG motif N rewires the G3BP1/2 interactome to disrupt stress granules.
Significantly, a peptide-based inhibitor disrupting the G3BP1/2-N interaction dampened SARS-CoV-2 infection showing that the study findings can be directly translated into novel specific antiviral reagents.
The study findings were published in the peer reviewed journal: Nature Communications.
https://www.nature.com/articles/s41467-021-26498-z
Typically, viruses invade our cells and turn them into virus factories.
The study team identified the way SARS-CoV-2 takes control of our cells and the team has found a possible way to inhibit the ravages of the virus.
Dr Ylva Ivarsson, a professor of biochemistry at Uppsala University, who coordinated the study told Thailand
Medical News, "This could lead to the further development of an inhibitor against COVID-19, although a lot of work remains to be done before we can say this with certainty."
The SARS-CoV-2 coronavirus, like all viruses, is an intracellular parasite. Like other viruses, SARS-CoV-2 needs to use the machinery of cells to enter them and cause them to produce more viruses.
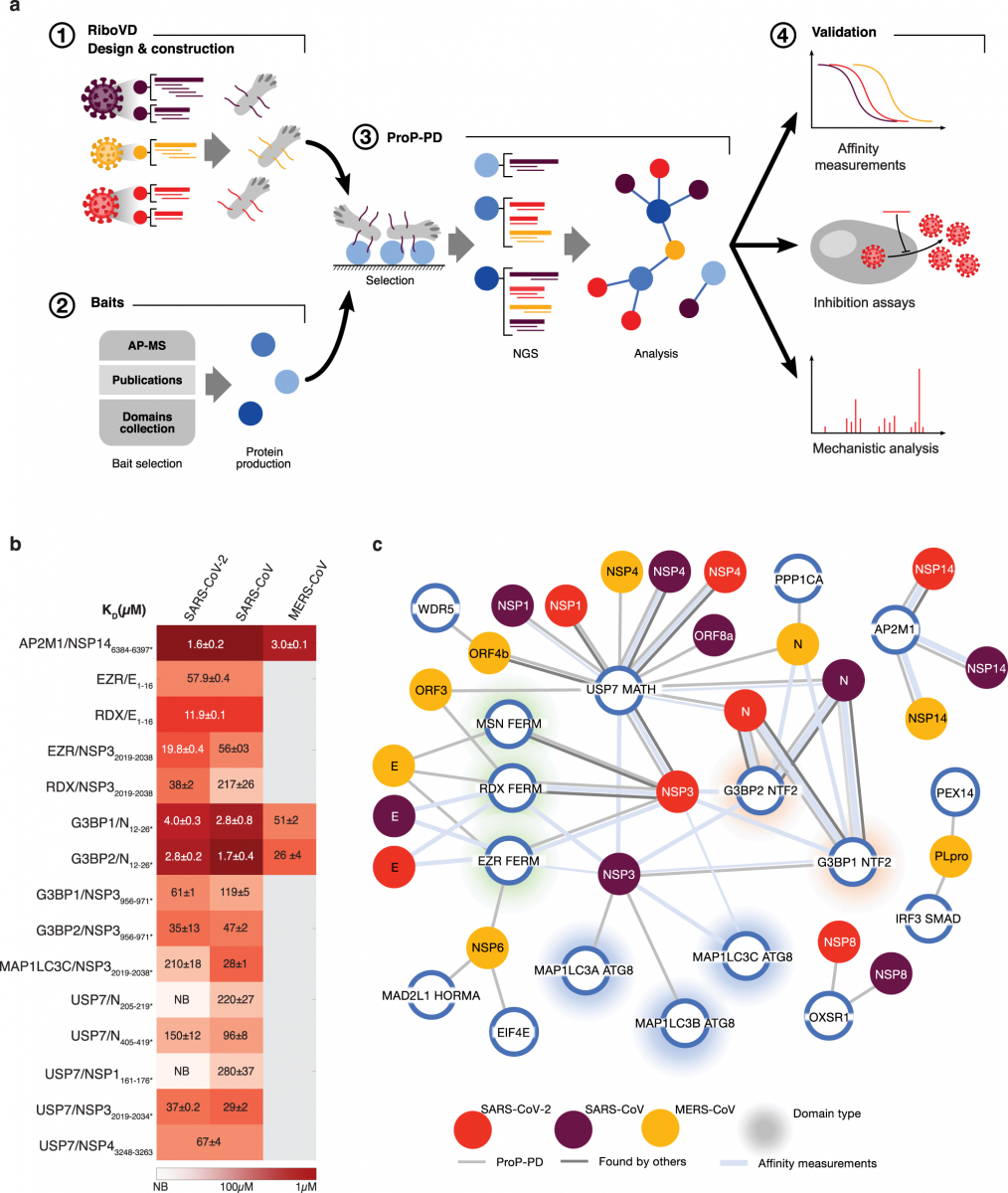 a An overview of the platform for identifying viral SLiMs binding to cellular host factors. b KD values for the interactions between indicated viral peptides and host proteins. c Network of SLiM mediated interactions between the indicated viral proteins from SARS-CoV-2 (red), SARS-CoV (purple), and MERS-CoV (yellow) and cellular host factors (blue circles). Light grey connecting line indicates interactions validated by affinity measurements, the weight of the
line represents the affinity of the interaction (thick, 1–10 μM; medium, 11–100 μM; thin, 101–500 μM). Dark grey lines indicate protein-protein interactions with additional evidence found in the other studies
a An overview of the platform for identifying viral SLiMs binding to cellular host factors. b KD values for the interactions between indicated viral peptides and host proteins. c Network of SLiM mediated interactions between the indicated viral proteins from SARS-CoV-2 (red), SARS-CoV (purple), and MERS-CoV (yellow) and cellular host factors (blue circles). Light grey connecting line indicates interactions validated by affinity measurements, the weight of the
line represents the affinity of the interaction (thick, 1–10 μM; medium, 11–100 μM; thin, 101–500 μM). Dark grey lines indicate protein-protein interactions with additional evidence found in the other studies
In order to produce more virus particles, the cell first needs to be induced to produce new genetic material, RNA, which is then wrapped in a protective coating and released from the cell to infect more cells. Though the virus cannot achieve this on its own, its proteins bind to human proteins and cause them to help produce viruses.
Planning to find key interactions that can be inhibited with existing or completely new medications, the study team worked hard since the start of the COVID-19 pandemic to determine which human proteins SARS-CoV-2 proteins bind to.
The study team developed and used a new method to map interactions on a large scale between human proteins and coronavirus proteins, which has provided valuable new information.
The study team used this information to show that blocking one of these interactions inhibited infection of human cells by SARS-CoV-2.
The study team confirmed the interaction between the viral protein and the human protein in a more complex system. This has enhanced understanding of what happens in the cell when the virus takes over the interaction.
The study findings could lead to the further development of an agent that inhibits COVID-19, although much work remains to be done before researchers can determine whether the inhibitor the study team found can be developed to treat the SARS-CoV-2 viral infection.
Dr Ivarsson added, "We have worked intensively on the project during the ongoing pandemic. It has been amazing how quickly research can progress when researchers from different universities and countries join forces and work towards the same goal."
Besides interactions with SARS-CoV-2 that cause COVID-19, the study team can also demonstrate many interactions with other coronavirus proteins, such as SARS-CoV and MERS-CoV and more common coronaviruses. This provides broad insight into how various coronaviruses differ and could ultimately help us to better prepare to deal with new types of viruses.
For the latest
SARS-CoV-2 research, keep on logging to Thailand Medical News.

 a An overview of the platform for identifying viral SLiMs binding to cellular host factors. b KD values for the interactions between indicated viral peptides and host proteins. c Network of SLiM mediated interactions between the indicated viral proteins from SARS-CoV-2 (red), SARS-CoV (purple), and MERS-CoV (yellow) and cellular host factors (blue circles). Light grey connecting line indicates interactions validated by affinity measurements, the weight of the
line represents the affinity of the interaction (thick, 1–10 μM; medium, 11–100 μM; thin, 101–500 μM). Dark grey lines indicate protein-protein interactions with additional evidence found in the other studies
a An overview of the platform for identifying viral SLiMs binding to cellular host factors. b KD values for the interactions between indicated viral peptides and host proteins. c Network of SLiM mediated interactions between the indicated viral proteins from SARS-CoV-2 (red), SARS-CoV (purple), and MERS-CoV (yellow) and cellular host factors (blue circles). Light grey connecting line indicates interactions validated by affinity measurements, the weight of the
line represents the affinity of the interaction (thick, 1–10 μM; medium, 11–100 μM; thin, 101–500 μM). Dark grey lines indicate protein-protein interactions with additional evidence found in the other studies