BREAKING! Study Discovers Presence Of Prion-Like Domains In SARS-CoV-2 That Affects Binding Affinity To ACE2! Every Variant Has Different Prion-Like Domain!
Source: SARS-CoV-2 Prion-Like Domains Feb 01, 2022 3 years, 2 months, 2 weeks, 5 days, 23 hours, 32 minutes ago
Researchers from the Human Microbiology Institute-New York, USA and Tetz Laboratories-New York, USA, have in a new study discovered the presence of prion-like domains in SARS-CoV-2 that affects binding affinity to ACE2. Further they also found that these prion-like domains vary in the various SARS-CoV-2 variants, contributing to their difference in binding affinity.

Prions are proteins that possess a unique conformational conversion, with the ability to rapidly shift between multiple conformations due to residue hydrophobicity and net sequence charge, and viral prion-like proteins are known as potential regulators of viral infections.
However, the
prion-like domains (PrD) in the SARS-CoV-2 proteome have not been analyzed.
The study team in this silico study, using the PLAAC algorithm, identified the presence of prion-like domains in the SARS-CoV-2 spike protein. Compared with other viruses, a striking difference was observed in the distribution of prion-like domains in the spike protein since SARS-CoV-2 is the only coronavirus with a prion-like domain found in the receptor-binding domain of the S1 region of the spike protein.
The presence and unique distribution of prion-like domains in the SARS-CoV-2 receptor-binding domains of the spike protein are particularly interesting since although the SARS-CoV-2 and SARS-CoV S proteins share the same host cell receptor, angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 demonstrates a 10- to 20-fold higher affinity for ACE2.
The study team identified prion-like domains in the α1 helix of the ACE2 receptor that interact with the viral receptor-binding domain of SARS-CoV-2.
The study team also found substantial differences in the prion-like domain of the S1 region of the spike protein across emerging variants including Omicron (B.1.1.529).
The study findings indicate that the identified PrDs in the SARS-CoV-2 receptor-binding domain (RBD) and ACE2 region that interact with RBD play important functional roles in viral adhesion and entry.
The study findings were published in the peer reviewed journal: Microorganisms.
https://www.mdpi.com/2076-2607/10/2/280/htm
The SARS-CoV-2 virus is a large, enveloped, single-stranded RNA virus that belongs to the family Coronaviridae of the genus Betacoronavirus (β-CoV).
 Analysis and comparison of mutations in the RBD of SARS-CoV-2 and SARS-CoV. The RBD of the SARS-CoV-2 (NCBI reference sequence: YP_009724390.1) spike protein was aligned with the closest related human βCoV, SARS-CoV (NCBI reference sequence: AYV99817.1). The PrDs of SARS-CoV-2 are red. Different residues are denoted by * beneath the consensus position. The amino acids asparagine (Q) and glutamine (N) in the PrDs of the SARS-CoV-2 RBD that differ from the amino acids in the SARS-CoV RBD are denoted by a red ** beneath the consensus position. Amino acids of the SARS-CoV-2 RBD that bind to ACE2 are marked with red boxes. RBD-receptor-binding dom
ain.
Analysis and comparison of mutations in the RBD of SARS-CoV-2 and SARS-CoV. The RBD of the SARS-CoV-2 (NCBI reference sequence: YP_009724390.1) spike protein was aligned with the closest related human βCoV, SARS-CoV (NCBI reference sequence: AYV99817.1). The PrDs of SARS-CoV-2 are red. Different residues are denoted by * beneath the consensus position. The amino acids asparagine (Q) and glutamine (N) in the PrDs of the SARS-CoV-2 RBD that differ from the amino acids in the SARS-CoV RBD are denoted by a red ** beneath the consensus position. Amino acids of the SARS-CoV-2 RBD that bind to ACE2 are marked with red boxes. RBD-receptor-binding dom
ain.
Aside from the SARS-CoV-2 virus, other members of the coronavirus family that have caused epidemics and claimed numerous lives are severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS). However, coronaviruses, such as common cold, human coronavirus OC43 (HCoV-OC43), and human coronavirus HKU1 (HCoV-HKU1) are non-life threatening.
It has been found that all these coronaviruses chiefly infect epithelial cells of the lungs; however, the clinical severity and pathogenesis differ between coronaviruses. For instance, pulmonary fibrosis and severe pneumonia are associated with SARS-CoV-2, SARS, and MERS, but HCoV-OC43 and HCoV-HKU1 do not cause these symptoms.
As with all the other β-CoVs, SARS-CoV-2 encodes four structural proteins, namely, the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The key determining factor of the host specificity of β-CoVs is the S protein, which is also involved with facilitating viral entry into the host cell.
The spike or S protein contains two large domains, i.e., the N-terminal S1 and C-terminal S2. S1 region contains the receptor-binding domain (RBD), which detects and binds to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell, whereas S2 promotes membrane fusion.
Past studies have shown that the genomic variability of S1 is higher than S2, i.e., the S2 region is highly conserved.
https://www.sciencedirect.com/science/article/pii/S0966842X15001316
https://www.nature.com/articles/s41541-020-0170-0
Numerous studies have shown that compared to SARS-CoV, SARS-CoV-2 has a higher binding affinity to ACE2 of the host.
https://www.science.org/doi/full/10.1126/science.abb2507
https://onlinelibrary.wiley.com/doi/full/10.1111/jcmm.15541
https://pubmed.ncbi.nlm.nih.gov/33806624/
https://pubmed.ncbi.nlm.nih.gov/33449359/
https://pubmed.ncbi.nlm.nih.gov/34571756/
Many SARS-CoV-2 variants have emerged over the course of the pandemic due to genomic mutations. Some of the SARS-CoV-2 variants have exhibited higher transmission rates, virulence, and shown the ability to evade immune responses induced via immunization or natural infection. These variants have been classified as variants of concern (VOC) and variants of interest (VOI) by the World Health Organization.
 Interactions between amino acids of PrDs and non-prion-like regions of SARS-CoV-2 RBD and ACE2. Amino acids Q498 and T500 from the PrD of the SARS-CoV2 RBD interact with Y41 and Q42 within the PrD of ACE2, while Q474, F486 and N501 from the PrD of the SARS-CoV-2 RBD bind to Q24, M82 and K343 from the non-PrD of ACE2. K417 and Y453 were the only amino acids of the SARS-CoV-2 RBD that were outside the viral PrD and bound to ACE2.
Interactions between amino acids of PrDs and non-prion-like regions of SARS-CoV-2 RBD and ACE2. Amino acids Q498 and T500 from the PrD of the SARS-CoV2 RBD interact with Y41 and Q42 within the PrD of ACE2, while Q474, F486 and N501 from the PrD of the SARS-CoV-2 RBD bind to Q24, M82 and K343 from the non-PrD of ACE2. K417 and Y453 were the only amino acids of the SARS-CoV-2 RBD that were outside the viral PrD and bound to ACE2.
The prion-like domains (PrDs) are highly associated with virus-host cell interactions. Although the molecular mechanisms behind prion formation are not clear, researchers found that the prion formation is driven by hydrophobicity, which is characterized by the presence of asparagine (Q)- and glutamine (N)-rich regions, and net sequence charge.
https://pubmed.ncbi.nlm.nih.gov/11050225/
https://pubmed.ncbi.nlm.nih.gov/23719919/
Numerous PrDs predictive algorithms are available, for example, prion-like amino acid composition (PLAAC) analysis. This algorithm allows the analysis of PrDs based on the hidden Markov model (HMM). To date, there is scarcity in research associated with the prionogenic properties of SARS-CoV-2.
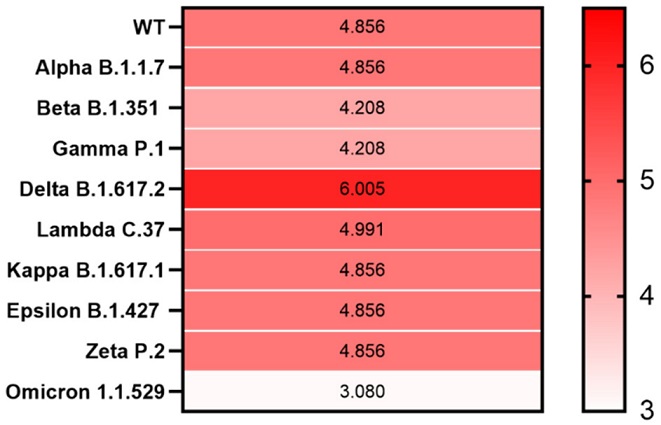 Heatmap showing PrD within the S protein in SARS-CoV-2 variants. The correlation between the LLR scores of the identified PrDs in the S protein across different SARS-CoV-2 variants is presented. Mean LLC scores of S protein are denoted using a color scale, ranging from white (minimum) to saturated red (maximum). Higher LLC scores indicate a higher possibility that the analyzed protein is a prion
Heatmap showing PrD within the S protein in SARS-CoV-2 variants. The correlation between the LLR scores of the identified PrDs in the S protein across different SARS-CoV-2 variants is presented. Mean LLC scores of S protein are denoted using a color scale, ranging from white (minimum) to saturated red (maximum). Higher LLC scores indicate a higher possibility that the analyzed protein is a prion.
The study team detected and analyzed viral PrDs in the S protein of SARS-CoV-2, which are the novel regulators of virion assembly.
The study team also compared them with the PrDs of other human-pathogenic β-CoVs. The study has been published in Microorganisms.
The team also determined PrDs in the S protein of the SARS-CoV-2 variants such as B.1.617.2 (Delta), B.1.1.529 (Omicron), B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.427 (Epsilon), B.1.617.1 (Kappa) and P.2 (Zeta).
The study findings of this research are in line with previous studies that analyzed the pathological function of prions in humans (e.g., Alzheimer’s and Parkinson’s diseases, and diabetes). Scientists have stated that protein misfolding plays an important part in both prokaryotes and eukaryotes.
The study team in the current research used the ratio of the high threshold of the PLAAC score for protein identification and only proteins with a high probability of prionogenic properties.
The study team observed that even though different β-CoV members contain PrDs in the S proteins, SARS-CoV-2 was the only coronavirus that contained PrD in the RBD of the S protein.
The study team discovered specific amino acids, i.e., Q474, N481, Q493, Q498, and N501, that allow the prionogenity of the SARS-CoV-2 RBD, which interacts within ACE2.
Past studies have investigated the atomic interaction of SARS-CoV-2 and ACE2 and detected several prion-like segments that exhibited pairwise interactions within the intrinsic disorder. The authors stated that as PrDs are only present in the RBD of SARS-CoV-2, their presence must benefit the virus.
https://pubmed.ncbi.nlm.nih.gov/33449359/
https://pubmed.ncbi.nlm.nih.gov/34571756/
https://pubmed.ncbi.nlm.nih.gov/11050225/
https://pubmed.ncbi.nlm.nih.gov/34142653/
The study findings provide proof of this concept, revealing that the presence of PrDs in the RBD of SARS-CoV-2 increases the affinity of the virus to bind to the ACE2 receptor of the host.
The study findings also revealed that among all the SARS-CoV-2 variants studied, the highest log-likelihood ratio scores were observed in the S protein of the SARS-CoV-2 Delta variant. This finding is significant because the Delta variant exhibits a high transmission rate. Additionally, this variant has been associated with higher mortality and hospitalization.
However, the emerging Omicron (B.1.1.529) variant, although known for its high transmissibility, for now, appears to be milder than previous strains and has the lowest LLR score among all SARS-CoV-2 variants.
The study team stated that the current study is the most complete evaluation of PrDs in the S protein of SARS-CoV-2 variants. However, more analysis associated with the PrD-containing proteins in SARS-CoV-2 is required to understand COVID-19 infection better.
The study findings have also provided new perceptions regarding the pathophysiology as well as novel targets that could be used to develop effective COVID-19 therapeutics and vaccines.
For more about
Prion-Like Domains in SARS-CoV-2, keep on logging to Thailand Medical News.

 Analysis and comparison of mutations in the RBD of SARS-CoV-2 and SARS-CoV. The RBD of the SARS-CoV-2 (NCBI reference sequence: YP_009724390.1) spike protein was aligned with the closest related human βCoV, SARS-CoV (NCBI reference sequence: AYV99817.1). The PrDs of SARS-CoV-2 are red. Different residues are denoted by * beneath the consensus position. The amino acids asparagine (Q) and glutamine (N) in the PrDs of the SARS-CoV-2 RBD that differ from the amino acids in the SARS-CoV RBD are denoted by a red ** beneath the consensus position. Amino acids of the SARS-CoV-2 RBD that bind to ACE2 are marked with red boxes. RBD-receptor-binding dom
ain.
Analysis and comparison of mutations in the RBD of SARS-CoV-2 and SARS-CoV. The RBD of the SARS-CoV-2 (NCBI reference sequence: YP_009724390.1) spike protein was aligned with the closest related human βCoV, SARS-CoV (NCBI reference sequence: AYV99817.1). The PrDs of SARS-CoV-2 are red. Different residues are denoted by * beneath the consensus position. The amino acids asparagine (Q) and glutamine (N) in the PrDs of the SARS-CoV-2 RBD that differ from the amino acids in the SARS-CoV RBD are denoted by a red ** beneath the consensus position. Amino acids of the SARS-CoV-2 RBD that bind to ACE2 are marked with red boxes. RBD-receptor-binding dom
ain. Interactions between amino acids of PrDs and non-prion-like regions of SARS-CoV-2 RBD and ACE2. Amino acids Q498 and T500 from the PrD of the SARS-CoV2 RBD interact with Y41 and Q42 within the PrD of ACE2, while Q474, F486 and N501 from the PrD of the SARS-CoV-2 RBD bind to Q24, M82 and K343 from the non-PrD of ACE2. K417 and Y453 were the only amino acids of the SARS-CoV-2 RBD that were outside the viral PrD and bound to ACE2.
Interactions between amino acids of PrDs and non-prion-like regions of SARS-CoV-2 RBD and ACE2. Amino acids Q498 and T500 from the PrD of the SARS-CoV2 RBD interact with Y41 and Q42 within the PrD of ACE2, while Q474, F486 and N501 from the PrD of the SARS-CoV-2 RBD bind to Q24, M82 and K343 from the non-PrD of ACE2. K417 and Y453 were the only amino acids of the SARS-CoV-2 RBD that were outside the viral PrD and bound to ACE2.