Chinese Study Finds That Human Bivalent COVID-19 Neutralizing Antibodies More Potent And Effective In Inhibiting SARS-CoV-2 Infections
Source: COVID-19 Neutralizing Antibodies Oct 15, 2020 4 years, 6 months, 1 week, 4 days, 7 hours, 22 minutes ago
COVID-19 Neutralizing Antibodies: A new study by researchers and immunologists from Westlake University, Zhejiang-China, Tsinghua University, Beijing-China and Southern University of Science and Technology, Shenzhen-China has found that human bivalent anti-spike antibodies are more effective in terms of viral neutralization and controlling a SARS-CoV-2 infection in humans.
.png)
The study team summed up their research, ”Neutralizing monoclonal antibodies (nAbs) to SARS-CoV-2represent promising candidates for clinical intervention against the COVID-19 disease. We isolated a large number of nAbs from SARS-CoV-2 infected individuals capable of disrupting proper interaction between the receptor binding domain (RBD) of the viral spike (S) protein and the receptor angiotensin converting enzyme 2 (ACE2). In order to understand the mechanism of these nAbs on neutralizing SARS-CoV-2 virus infections, we performed cryo-EM analysis and here report cryo-EM structures of the ten most potent nAbs in their native full-length IgG or Fab forms bound to the trimeric S protein of SARS-CoV-2.
The bivalent binding of the full-length IgG is found to associate with more RBD in the "up" conformation than the monovalent binding of Fab, perhaps contributing to the enhanced neutralizing activity of IgG and triggering more shedding of the S1 subunit from the S protein. Comparison of large number of nAbs identified common and unique structural features associated with their potent neutralizing activities. This work provides structural basis for further understanding the mechanism of nAbs, especially through revealing the bivalent binding and their correlation with more potent neutralization and the shedding of S1 subunit.”
The research findings were published on a preprint server and have yet to be peer reviewed.
https://www.biorxiv.org/content/10.1101/2020.10.13.336800v1
The current COVID-19 crisis shows no signs of fading and has prompted more efforts to develop effective antiviral therapeutics and vaccines against the SARS-CoV-2 virus.). The new study on the effective viral neutralization by bivalent anti-spike antibodies could promote the development of better therapeutics and vaccines.
The SARS-CoV-2 virus has about 80% of the same sequences as the earlier SARS-CoV. Both attach to the same host receptor, angiotensin-converting enzyme 2 (ACE2), via the S1 subunit of the viral spike protein. This is followed by the proteolytic cleavage of the spike protein at the S1/S2 interface, with the S2 undergoing marked conformation change shedding the S1 subunit to give rise to the post-fusion form of the spike. This facilitates viral entry into the cell.
Importantly the S1 subunit has the receptor-binding domain (RBD), which is targeted by many natural anti-spike antibodies as well as many vaccines under development. This is the part of the S1 subunit that directly attaches to the receptor.
It has been found that in the prefusion state, the spike structure shows more significant changes in conformation in the S1 region, and this is especially the case with the RBD. This can have either the ‘up’ or ‘down’ conformations, and only the former can attach to the ACE2 receptor.&l
t;br />
The research aims at understanding how neutralizing antibodies (nAbs), which are typically bivalent, bind to the trimeric spike protein. The researchers used ten cryo-EM structures of the spike-nAb (IgG) complex, with either full-length nAb or Fab fragment, or both.
The study team identified ten nAbs from plasma obtained from recovered COVID-19 patients. All were strongly neutralizing, competing with the ACE2 receptor to bind RBD. They also found that while some nAbs readily induced S1 subunit shedding, others did not.
Significantly, by structural analysis, the team found three patterns to the nAb-spike complexes. The first included four of the ten nAbs, which bound two ‘up’ RBDs. The second pattern, shown by three nAbs, had either two or three RBDs bound in the ‘up’ conformation to the antibody.
The third pattern was shown by the most potent nAb examined here, which bound one RBD in the ‘up’ and two in the ‘down’ conformation, all three RBDs sharing the same binding interface.
Past studies have suggested that bivalent nAbs are more potent compared to Fabs in viruses like Dengue and rhinoviruses. The researchers, therefore, imaged the Fab-Spike complex separately. They found that this showed only two RBDs bound to the nAb, both in the ‘up’ conformation. With the IgG-spike complex, two or three RBDs bind to the nAbs in “up” conformation.
However despite different binding patterns, all full-length antibodies showed bivalent binding of the viral spike protein.
The study team also found a difference in the orientation of the RBDs bound to the Fab and the full-length IgG. The half-maximal inhibitory concentration (IC50) values were 700 times lower with the full-length antibody than the Fab form for two of three tested antibodies, while with the third, it was over 3,000 times less.
Interestingly S1 subunit shedding was found to be at over 80% of the S protein at an incubation time of 120 minutes, with one of the IgG nAbs, while the Fab form of the same antibody caused hardly any shedding. Other nAbs showed mild shedding only.
It was also found that the full-length IgG nAbs were thus able to bind more strongly and had more potent neutralization capacity, as well as inducing more significant S1 shedding, compared to the Fab fragment binding alone to the spike RBD.
Previous findings showed that S1 shedding has been shown to be a possible correlate of neutralizing power since cross-reactive nAbs to SARS-CoV and SARS-CoV-2 are typically directed against RBD, prevent ACE2 binding, and promote S1 shedding. However, since the native spike protein exists as a homotrimer, it is unknown whether all the S1 subunits have to be shed to allow the spike protein to change from the prefusion to the post-fusion form.
The detailed structure of the Fab and the scFv was constructed for 8/10 and the remaining 2/10 antibodies, respectively. This showed three groups of antibodies concerning the epitopes and the angles at which they bound to the RBD.
It was found that in the first group of 7 nAbs, there was a high overlap between the receptor-binding motif (RBM) on the RBD and the epitopes. These antibodies recognized 8-15 of the 17 epitope residues that mediate binding to the ACE2 receptor. The angles at which the antibodies make contact with the RBD are also comparable between nAbs in this group.
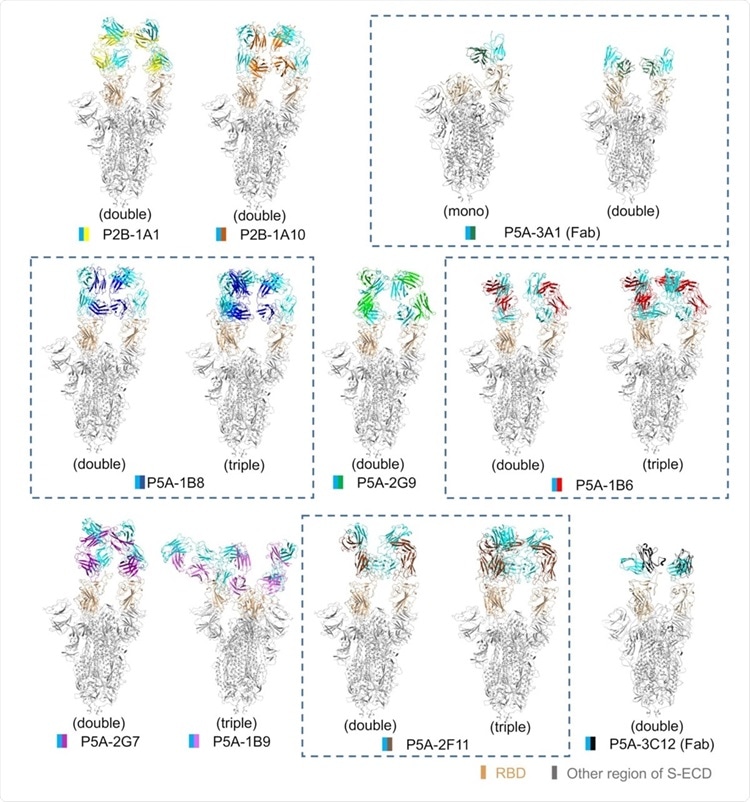 All solved structures of nAbs in complex with S protein. The domain-colored models of all complex are shown here. The structures containing different numbers of the same nAb are boxed with blue dash line. The structures are labeled according to the number of RBD bound with nAb as mono (1 RBD), double (2 RBDs) or triple (3 RBDs) binding, respectively.
All solved structures of nAbs in complex with S protein. The domain-colored models of all complex are shown here. The structures containing different numbers of the same nAb are boxed with blue dash line. The structures are labeled according to the number of RBD bound with nAb as mono (1 RBD), double (2 RBDs) or triple (3 RBDs) binding, respectively.
This study confirms that of the 17 anti- SARS-CoV-2 nAbs in the Protein Data Bank (PDB) database, group 1 nAbs are the most common.
The group 2 contains only one nAb, the most potent nAb tested in this study against both the wildtype and the pseudo-SARS-CoV-2. This antibody shows pattern 3 binding to the RBD. Its direction of the RBD approach is different from that of group 1 nAbs.
The group 3 nAbs bind to the remote loop of the RBM and have little overlap with the RBM alone, with an unique binding mechanism.
Significantly of the three subgroups in group 1, the first two induce the shedding of over 77% of S1, but this ability is much weaker, at ~44% to ~57%, with subgroup 3 and groups 2 and 3. The researchers feel, “The S1 shedding ability of the nAbs may be facilitated by the large overlap with RBM.”
Importantly subgroup 3 of group 1 nAbs shows rotation of the longitudinal axis of the Fab compared with the antibodies in subgroup 1 and 2. It is possible that only if these antibodies can achieve a specific angle of binding with the RBD can they cause shedding of the S1 subunit.
The presence of the Fc region again in the antibody-virus complex can increase the immune response by allowing T cell binding via their Fc receptors.
Implications
Hence this showed that antibody binding in the full-length form uses a different mode and causes more RBDs to flip to the ‘up’ conformation than with the Fab form. The former involves bivalent and the latter monovalent binding. Bivalent binding is more effective in inhibiting viral entry and is associated with more significant S1 subunit shedding.
Previously, the same study team showed the dimeric nature of the ACE2 receptor. If so, each of the ACE2 molecules can bind one of the RBDs in the S trimer, thus inducing the bound RBD to flip ‘up’. Steric hindrance probably prevents the binding of the two ACE2 molecules in the dimer to different RBDs of the S trimer at the same time, and thus to flip them ‘up.’ This would require the binding of more than one ACE2 receptor.
Though many past studies showed many complex spike-nAb structures, most of them used the Fab form of the antibody, particularly if X-ray crystallization was used for the structural visualization.
To date, there are 17 structures involving anti-SARS-CoV-2 nAbs in the Protein Data Bank (PDB) database. The group 1 nAbs, which have the largest overlap between the epitope and RBM, not only are most popular in this work, but have many similar nAbs in the PDB database, including B38 (PDB code: 7BZ5)(26), CB6 (PDB code: 7C01)(23), C105 (PDB code: 6XCN)(29), CV30 (PDB code: 6XE1)(30), CC12.1 (PDB code: 6XC2)(31), CC12.3 (PDB code: 6XC4)(31), COVA2-04 (PDB code: 7JMO)(32) and COVA2-39 (PDB code: 7JMP)(32). Interestingly, the
P2B-1A10,
P5A-3A1, P5A1B8, B38, C105, CV30, CC12.1, CC12.3, COVA2-04 and COVA2-39 are all belong to the IGHV3-53 gene family.
The group 2 nAb P5A-1B9 has the highest inhibitory activities against the cell infection of both pseudo and live SARS-CoV-2. Structure of the complex of P5A-1B9 with S protein shows that it can bind to RBD in both up and down conformation, similar to the nAb BD-368-2, suggesting a common mechanism behind these nAbs of very high potency. The epitope of P5A-1B9 is only 6-residue overlapped with RBM, and is similar to P2B-2F6 (PDB code: 7BWJ)(17), Fab2-4 (PDB code: 6XEY)(34) and BD23 (PDB code: 7BYR)(35), but they have different gene family. P2B-2F6 and Fab2-4 have different approaching direction between the nAbs and RBD to the group 1 nAbs. The group 3 nAbs, P5A-2F11 and P5A-3C12, only bind to the remote loop of RBM and there is no published nAb of the similar binding mode
The research findings show that the full-length IgG is more physiological and should be preferred for structural analysis to shed light on the actual structure and function of nAbs targeting SARS-CoV-2.
For more on
COVID-19 Neutralizing Antibodies, keep on logging to Thailand Medical News.
.png)
