Coronavirus News: Washington University Study Identifies SARS-CoV-2 Mutations That Escape Monoclonal Antibodies And Polyclonal Immune Sera
Source: Coronavirus News Feb 09, 2021 4 years, 2 months, 2 weeks, 3 days, 15 hours, 2 minutes ago
Coronavirus News: Researchers from the School of Medicine at Washington University have in a new study identified SARS-CoV-2 mutations that escape monoclonal antibodies and polyclonal immune sera.
The neutralizing antibodies against the SARS-CoV-2 spike (S) protein are a goal of COVID-19 vaccines and have received emergency use authorization as therapeutics.
However, viral escape mutants could compromise vaccine efficacy. To define immune-selected mutations in the S protein, the study team exposed a VSV-eGFP-SARS-CoV-2-S chimeric virus, in which the VSV glycoprotein is replaced with the S protein, to 19 neutralizing monoclonal antibodies (mAbs) against the receptor-binding domain (RBD) and generated 50 different escape mutants.
Interestingly each mAb had a unique resistance profile, although many shared residues within an epitope of the RBD. Some variants (e.g., S477N) were resistant to neutralization by multiple mAbs, whereas others (e.g., E484K) escaped neutralization by convalescent sera.
Furthermore sequential selection identified mutants that escape neutralization by antibody cocktails. Comparing these antibody-mediated mutations with sequence variation in circulating SARS-CoV-2 revealed substitutions that may attenuate neutralizing immune responses in some humans and thus warrants further investigation.
The study findings are published in the peer reviewed journal:
Cell Host & Microbe.
https://www.sciencedirect.com/science/article/pii/S1931312821000445#!
The current COVID-19 pandemic shows no signs of ending soon, the definitive way out are the achievement of population immunity by either vaccination or natural infection. With the unacceptably high mortality rates associated with the latter route, the development of effective vaccines that can induce neutralizing antibodies against the disease’s causative agent-SARS-CoV-2 coronavirus is of dire urgency.
Identified therapeutic antibodies and those elicited by current vaccines are effective in neutralizing viral antigens belonging to the spike protein of SARS-CoV-2.
But as new variants emerge, understanding the epitopes to which these protective antibodies bind is an important area of study.
Importantly it is also necessary to explore the development of natural resistance to neutralization because of these variations.
This new study describes the experimental selection of
SARS-CoV-2 mutants associated with resistance to antibodies, with important implications for vaccine development and therapeutic antibody use.
Typically in any RNA virus, there are multiple variant genome sequences, forming a swarm or quasispecies, around a core consensus genome. When neutralizing antibodies or drugs act on the virus, some variants will be selected because of their resistance to neutralization. This leads to the emergence of resistant variants, provided these have high fitness.
It should be noted that the coronaviruses&nbs
p;have a relatively low mutation rate among RNA viruses. This is because of the presence of a proofreading enzyme to correct random changes in the RNA genome introduced by the viral RNA-dependent RNA polymerases (RdRp).
The SARS-CoV-2 spike gene however still shows over 4,000 mutations, which have caused over 1,200 amino acid substitutions. Of these, almost 190 are in the receptor-binding domain (RBD). The presence of a large number of viral variants in circulation indicates the retention of viral fitness.
These non-synonymous mutations may persist due to host adaptation, selection of escape mutations by neutralizing antibodies formed during natural infection, and perhaps the reinfection of individuals with weakened immunity.
The study team utilized a panel of 19 antibodies to identify escape mutations.
The team also used a vesicular stomatitis virus (VSV) expressing SARS-CoV-2 spike, denoted by VSV-SARS-CoV-2. This replicating virus can form high titers while engaging the human angiotensin-converting enzyme 2 (ACE2) receptor. It is also subject to neutralization by anti-spike monoclonal antibodies.
Upon treating with neutralizing antibodies, the study team sequenced plaque-forming variants. These isolates were then allowed to infect cells in the presence and absence of antibodies to confirm that they were resistant.
The study team was able to isolate 50 escape mutants with multiple substitutions in the RBD, often within the region of direct contact with ACE2.
From 19 different mAbs that neutralize SARS-CoV-2, the team isolated 50 viral mutants that escape neutralization. Selection of escape mutants was facilitated by the use of VSV-SARS-CoV-2, which they previously validated as an effective mimic of SARS-CoV-2 S protein-mediated infection. The mAbs were obtained following immunization with soluble RBD, and although some mice received a boost with stabilized S ectodomain protein, all escape substitutions map within the RBD.
Multiple different mAbs led to resistance substitutions at K444, G446, N450, L452, S477, T478, P479, E484, F486, and P499, suggesting that they comprise major antigenic sites within the RBD. In earlier work, substitutions at residues K444, N450, E484, and F486 were identified using two antibodies in clinical developmen, and a separate study using three different antibodies defined resistance substitutions at R346, N440, E484, F490, and Q493.
These findings suggest that with many neutralizing antibodies that bind to the side of the RBD, the mechanism of action is by allosteric interference with spike-ACE2 binding. Another possible mechanism is by inhibiting the interactions of the virus with other attachment factors.
Significantly for mutations at the RBD base, allosteric inhibition of the ‘up’ conformation required for its binding to the ACE2 receptor may contribute to the resistance.
The study team also found that several mutations conferred resistance to more than one antibody. When multiple substitutions with different side chains were possible for a given residue, each of these changes showed a unique pattern of resistance.
Alarmingly S477N and S477G conferred resistance to all the antibodies used.
The study team recommends further study of the S477N mutation, a fairly common spike variant, because of its broad resistance to the panel of monoclonals studied here.
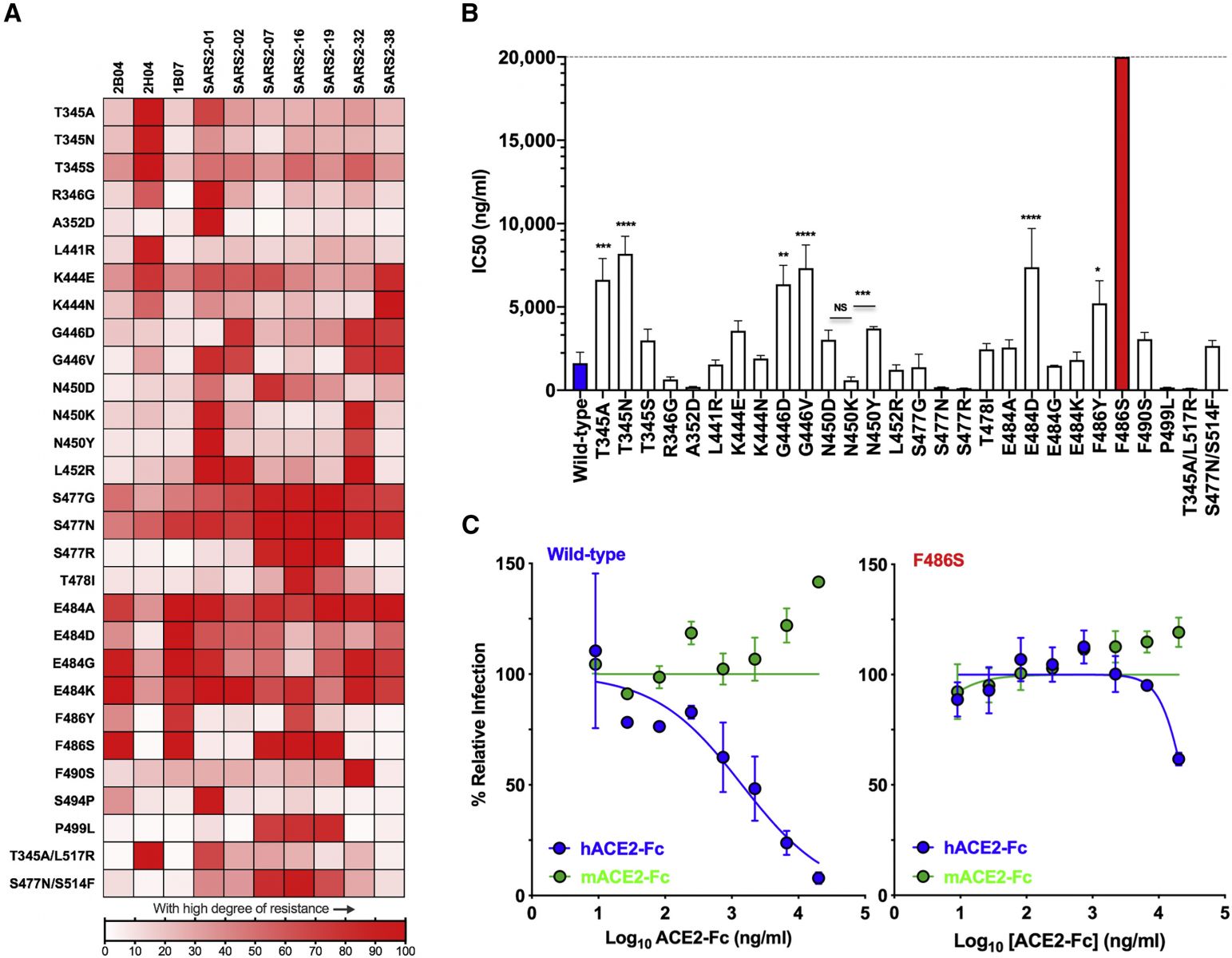
Map of cross-neutralizing activity of VSV-SARS-CoV-2 mutants and neutralization potency of hACE2 decoy receptors against each VSV-SARS-CoV-2 mutant. (A) Neutralization of VSV-SARS-CoV-2 mutants was evaluated by plaque assays. Degree of resistance was defined as percentage by expressing the number of plaques formed by each mutant in the presence versus absence of antibody and is represented as a heatmap from white (low degree of resistance) to red (high degree of resistance). (B) Neutralization assay of VSV-SARS-CoV-2 mutants in the presence of hACE2-Fc. Virus was incubated with mACE2 or hACE2 at concentrations ranging from 9 ng/mL to 20 μg/mL for 1 h a 37°C, and cells were scored for infection at 7.5 h post-inoculation by automated microscopy. IC50 values were calculated for each virus-hACE2 combination from three independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; one-way ANOVA with Dunnett’s post-test; error bars indicate SEM). (C) Representative neutralization curves of wild-type and F486S mutant VSV-SARS-CoV-2 with hACE2-Fc and mACE2-Fc. Error bars represent the SEM. Data are representative of three independent experiments.
It should also be noted however that this mutation was still susceptible to neutralization by human immune sera, indicating that this epitope is not immunodominant in humans.
Just as alarming, the E484 substitution by any of four possible residues was associated with escape from neutralization by four polyclonal human sera. One of the four sera was susceptible to escape only by a change at this position. Substitutions at E484 led to an increase in infectivity even in the presence of multiple sera.
Numerous other substitutions allowed escape from inhibition by two or three sera.
The F486S mutation was associated with escape from inhibition by multiple antibodies, as well as by soluble recombinant human ACE2, which was expected to act as a decoy for the virus, preventing its binding to the host cell ACE2 receptor.
The study team compared the 50 resistant variants with the clinical isolates and found that some variants already in circulation are resistant to both monoclonals and polyclonal antibodies.
Corresponding author Dr Sean P.J. Whelan from the Department of Molecular Microbiology, School of Medicine, Washington University in St. Louis told Thailand Medical News, “Neutralizing mAbs against RBD can select for variants or changes at positions that already exist within the human population.”
The study team was also able to demonstrate that the emergence of such escape mutants could be slowed, if not completely blocked, by using cocktails of monoclonal antibodies. Such combinations bind to different epitopes on the spike protein. However, if circulating strains already contain escape mutations to one of the antibodies in the cocktail, additional escape mutations may be selected.
The research findings show that several escape mutants, such as S477N, are resistant to neutralization by multiple monoclonal antibodies. Others like E484K show escape from neutralization by human immune sera. The latter finding indicates that the polyclonal neutralizing response in some convalescent patients, at least, is due to antibodies that bind only a few epitopes.
Also a good number of the escape mutants, such as E484, identified here show substitutions at residues where circulating isolates already display mutations.
These findings imply that escape variants can emerge in the presence of such a limited polyclonal response. This would limit the usefulness of vaccines that induce these neutralizing antibodies, indicating the need to increase the spectrum of neutralizing antibody responses.
Another implication is that effective therapeutic antibodies will probably be required in cocktail form, since escape mutations are so easily selected and are already present at high frequency in clinical isolates. An accurate knowledge of the association of different residues with resistance to each monoclonal antibody will help select the right combinations, avoiding overlapping escape mutations.
It should however be noted that resistance to such cocktails could still emerge via sequential escape, when a variant that escapes neutralization by one monoclonal subsequently acquires an escape mutation for another monoclonal. This process was demonstrated in the current study, and will help identify such mutants.
For the latest
Coronavirus News, keep on logging to Thailand Medical News.


