COVID-19 Genomics: Swiss Researchers Discover How SARS-CoV-2 NSP1 Protein Inhibits Host Translation. Findings Has Implications For Vaccines And Drugs
Source: COVID-19 Genomics Jul 09, 2020 4 years, 9 months, 3 weeks, 4 hours, 49 minutes ago
COVID-19 Genomics: Swiss Researchers from the Institute of Molecular Biology and Biophysics-Zurich, University of Bern and Institute of Virology and Immunology-Bern have discovered the detailed functional process by which the SARS-CoV-2 coronavirus NSP1 Protein Inhibits Host Translation.
.jpg)
Their study published in a preprint server shows that the 5’ end of the RNA molecule is capable of inhibiting the binding of mRNA to the ribosomal 40S subunit, thus preventing translation.
https://www.biorxiv.org/content/10.1101/2020.07.07.191676v1
Typically when a virus enters a host cell, the genomic RNA is translated by the cellular machinery to produce non-structural proteins (nsps). These are required to allow viral infection and viral mRNA synthesis.
The SARS-Cov-2 coronavirus is observed to have adapted to its human hosts by a slew of mechanisms to achieve these ends. Some of these are inhibition of protein synthesis by the host cell, as well as splitting mRNAs produced by the host cell. In the SARS-CoV, the host shutoff protein has been observed, which is a confounding factor called nsp1. Encoded by the first gene at the 5’ end, this is expressed first of all and is intended to close down the expression of host proteins, including immune proteins, immediately after the host cell is entered and infected.
The detailed examination of this molecule shows that while the N-terminal domain is structured, the C-terminal domain is flexibly disordered. By acting against the expression of type I interferon and antiviral molecules, nsp1 is a factor that possibly increases the virulence of the virus, and could be the basis for a live attenuated vaccine.
The Swiss research aimed to understand how nsp1 inhibits translation at the molecular level, how this can be suppressed by key mutations, and how it enhances the translation of reporters that contain full-length UTRs of the virus. This may explain how the nsp1 can suppress host translation but allow viral mRNAs to be translated.
The study team found that when the bacterially produced viral nsp1 was added to ribosomal particles, they found that it binds to both 40S and 80S complexes. CryoEM of the pooled ribosomal complexes showed that a preinitiation complex (PIC) was formed comprising several proteins, including the initiation factor elF3 core and initiator tRNA. Surprisingly, there was a density in the entrance channel of the mRNA, which was not due to the mRNA.
The researchers found that nsp1 binds only to 40S ribosomal subunits. This complex was therefore assembled and its structure examined by cryoEM. This revealed that the density was the C-terminal domain of nsp1.
Detailed docking analysis of the resulting molecular model into the 43S molecular map obtained earlier showed that the C-terminal end of this nsp1 is also found in association with the PIC. At high resolution, it was apparent that the C-terminal portion was within the mRNA entrance channel. This agrees with the findings in the 40S-nsp1 complex.
It was found that the nsp1 protein folded at the C-terminal part to form a pair of helices that engaged to the h18 of the 18S mRNA and to proteins uS3 of the head, and to uS5 and eS30, in the body, respect
ively. These form a network of interactions through multiple side chains.
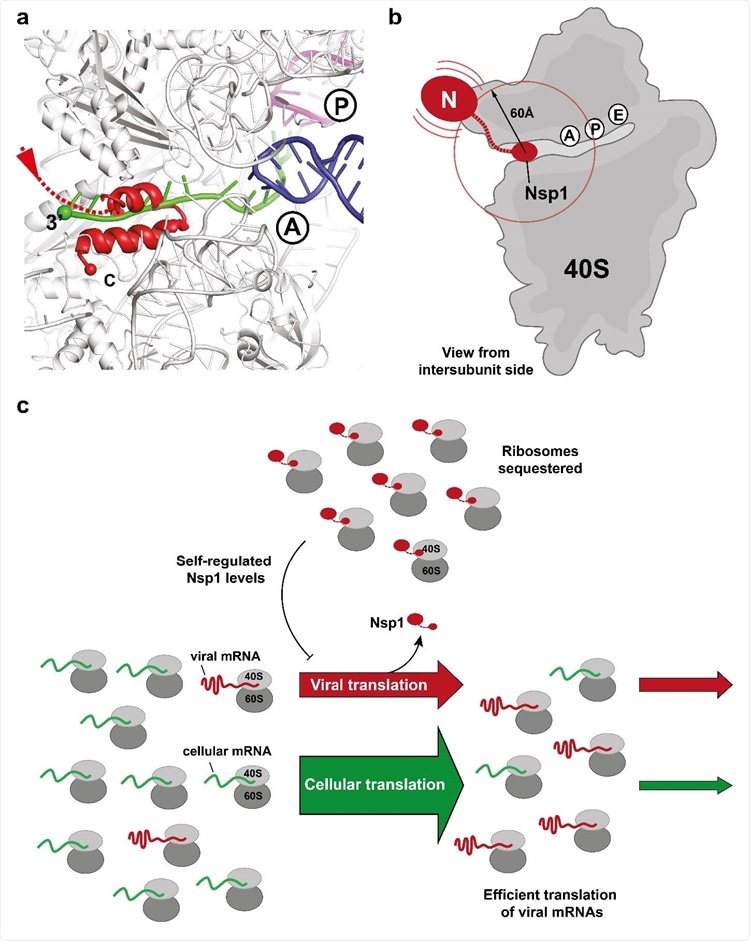 Binding of the C-terminal domain of SARS-CoV-2 Nsp1 to ribosomal mRNA channel prevents classical mRNA binding by sterical hindrance. (a) Superposition of canonically bound mRNA (green), A- (blue) and P-site (purple) tRNAs (pdb 6HCJ) reveals that Nsp1 (red) prevents classical binding of the mRNA at the entry site due to blockage. (b) Nsp1 binds via its C-terminus in proximity of the 40S mRNA entry site. Due to the flexible linker, the N-terminal domain can sample an area of ~60 Å around its attachment point (circle). (c) Model for translation inhibition by Nsp1. Upon viral infection and translation of viral genomic mRNA, Nsp1 acts as a translation inhibitor reducing the pool of ribosomes that can engage in translation. Under such ribosome-limiting conditions, viral mRNAs are translated with high efficiency.
Binding of the C-terminal domain of SARS-CoV-2 Nsp1 to ribosomal mRNA channel prevents classical mRNA binding by sterical hindrance. (a) Superposition of canonically bound mRNA (green), A- (blue) and P-site (purple) tRNAs (pdb 6HCJ) reveals that Nsp1 (red) prevents classical binding of the mRNA at the entry site due to blockage. (b) Nsp1 binds via its C-terminus in proximity of the 40S mRNA entry site. Due to the flexible linker, the N-terminal domain can sample an area of ~60 Å around its attachment point (circle). (c) Model for translation inhibition by Nsp1. Upon viral infection and translation of viral genomic mRNA, Nsp1 acts as a translation inhibitor reducing the pool of ribosomes that can engage in translation. Under such ribosome-limiting conditions, viral mRNAs are translated with high efficiency.
In both complexes, nsp1 binds to the mRNA entrance channel present in the 40S subunit. Here it overlaps with the mRNA even if the latter is wholly accommodated, leading to the inhibition of mRNA by nsp1 binding.
Dr Katharina Schubert, one of the co-researchers from Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich told Thailand Medical News, “Nsp1 is tightly bound to the 40S subunit through anchoring of its C-terminal helices to the mRNA channel while the N-terminal domain can sample space in the radius of approximately 60 Å from its attachment point.”
The study team also found that nsp1 almost completely inhibits translation of a luciferase reporter mRNA gene in a dose-dependent manner. Now, introducing some key mutations in helix 1 and 2, as well as in the KH motif, they found that this inhibition was abolished even at sixfold concentrations. They also failed to bind 40S ribosomal subunits, which indicate that the nsp1 has an affinity for binding the ribosome because of the C-terminal domain.
The researchers say this mechanism of inhibition may be found only in this virus and closely related beta-coronaviruses since only these have a conserved sufficiently long C-terminal that can bind the mRNA.
The team also analyzed how the translation of reporter mRNA with viral 5’ UTRS was different from cellular 5’ UTRs, and how nsp1 affected both. They found that the former was five times more efficient, but both were inhibited at varying concentrations of nsp1. This is an important finding since viral RNA is translated in competition with cellular mRNA.
On the whole, nsp1 is a general translation inhibitor, acting at the point of initiation. The mechanism is by sterically hindering the binding of mRNA by blocking the entrance of the mRNA channel. Further study is needed to understand how ribosomes are recruited to enable the viral mRNA to be efficiently translated.
One possible explanation might be that the potent inhibitory action of nsp1 leads to the tight binding of ribosomes and reduces the number of available ribosomes. In this restricted state, the ribosome will switch to the more efficient viral mRNA translation, because of the higher output achieved.
In order to enable this, the virus keeps nsp1 levels below the concentration that is required to inhibit viral mRNA translation, but enough to suppress the initiation from cellular mRNA.
Dr Schubert added, “Through this mechanism, we propose that Nsp1 would be able to inhibit global cellular translation particularly for mRNAs responsible for the host innate immune response, while the remaining ribosomes would still be able to translate viral mRNAs with high efficiency.”
It was observed that as the levels of viral mRNAs increase, up to 50% of total RNA in the cell, they will shift more and more of the translation machinery towards viral proteins.
Significantly, by identifying this key domain that interacts with ribosomes to control the cell’s response to the infection, attenuated vaccine strains will be easier to build.
Furthermore, these results provide an excellent basis for structure-based experiments aimed at investigating Nsp1 function in vivo by using viral model systems.
For more on
COVID-19 Genomics, keep on logging to Thailand Medical News.
.jpg)
