COVID-19 Immunology: Study Shows Human Gut Unique Immune Response To SARS-CoV-2 And Proviral Mechanisms To Shutdown IFN-Mediated Signaling
Source: COVID-19 Immunology Oct 29, 2020 5 years, 1 month, 2 weeks, 6 days, 9 hours, 4 minutes ago
COVID-19 Immunology: Researchers from the European Molecular Biology Laboratory (EMBL)-Germany, Heidelberg University-Germany, German Cancer Research Center-Heidelberg, Stanford University School of Medicine, Stanford Genome Technology Center and the University of California-San Diego have in a new study explored if the innate immune response triggered in the human gut to combat viral infection is similar or distinct compared to the one triggered in other organs.
.jpg)
The study team also compared the responses in infected and bystander cells. They used human ileum- and colon-derived organoids as a non-transformed culture model. The intestinal "mini-gut" organoids are an excellent model to study SARS-CoV-2 infection of the gastrointestinal tract.
This study of the immune response in the human gut upon SARS-CoV-2 infection revealed a robust proinflammatory response ie a cell type-specific or a tissue-specific regulation of interferon-mediated signaling during SARS-CoV-2 infection.
The study team identified a subpopulation of enterocytes as the prime target of SARS-CoV-2 and, interestingly, found the lack of positive correlation between susceptibility to infection and the expression of ACE2. Infected cells activated strong proinflammatory programs and produced interferon, while expression of interferon-stimulated genes was limited to bystander cells due to SARS-CoV-2 suppressing the autocrine action of interferon. These findings reveal that SARS-CoV-2 curtails the immune response and highlights the gut as a proinflammatory reservoir that should be considered to fully understand SARS-CoV-2 pathogenesis.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2020.10.21.348854v1
Many COVID-19 patients display gastroenteritis symptoms and there is growing evidence that the intestinal epithelium can be infected by SARS-CoV-2.
To date, little is known about the importance of the enteric phase of SARS-CoV-2 - both for the viral lifecycle and for the development of COVID-19-associated pathologies. It is not clear if the symptoms seen in patients are associated with the direct replication of SARS-CoV-2 in the gastrointestinal (GI) tract or are a consequence of the strong proinflammatory response.
Although It is understood that SARS-CoV-2 replicates in human intestinal epithelial cells; the specific cell-types that become infected are not well defined.
Utilizing single-cell transcriptomics approaches (scRNAseq) and targeted scRNAseq, the authors identify a subpopulation of enterocytes (namely, immature enterocytes 2) as the most susceptible to SARS-CoV-2 infection.
Significantly an important observation in this study is the lack of correlation between ACE2 expression level and the copy numbers of the SARS-CoV-2 genome in the cell. This implies that ACE2 expression cannot be used for conjectures on infectibility.
Hence the study team emphasizes that the investigation of SARS-CoV-2 tropism must be carried based on biological validation of infection and should not be done solely on the analysis of transcriptional profiles of individual cells or tissues.
/>
However since they found that TMPRSS2 (type II transmembrane serine protease 2) expression levels were associated with the SARS-CoV-2 genome copy numbers in human intestinal epithelial cells, they speculate that TMPRSS2 plays an essential role in the SARS-CoV-2 cell tropism, more so than the role of ACE2.
The study team also found that upon infection, ACE2 levels decrease both in infected and bystander hIECs (human intestinal epithelial cells). These show that the observed regulation of ACE2 upon infection might be tissue-specific and time-dependent.
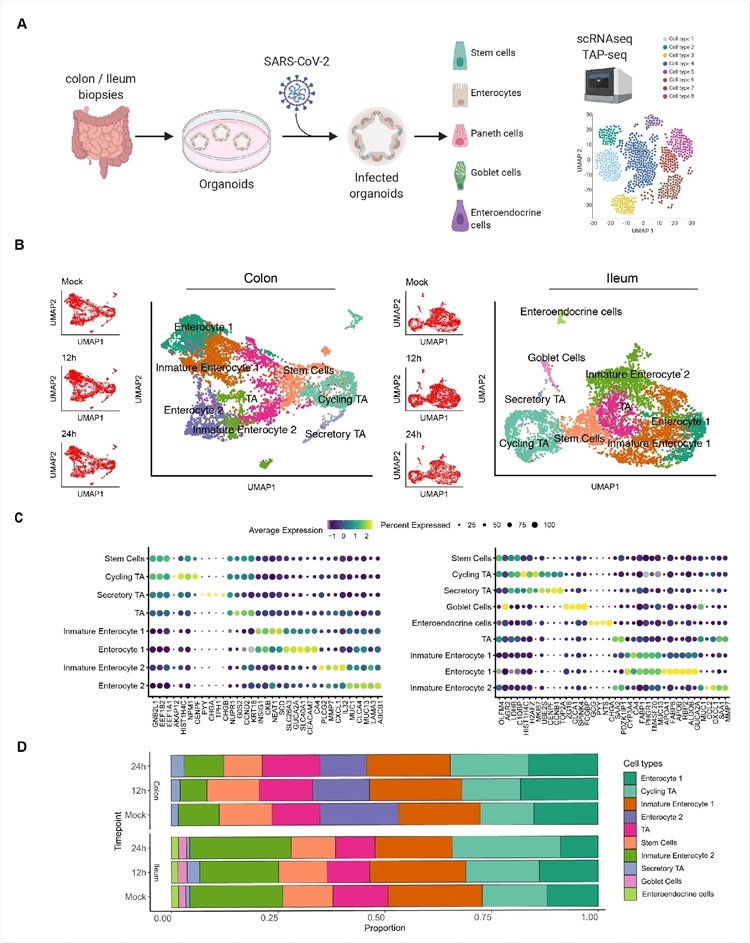
Single cell sequencing of SARS-CoV-2 infected colon- and ileum-derived human organoids. A. Schematic representation of the experimental workflow. 625 B. Uniform manifold approximation and projection (UMAP) embedding of single-cell RNA-Seq data from mock and SARS-CoV-2 infected colon-derived (left panels) and ileum-derived (right panels) organoids colored according to the cell type. Small insets represent the UMAP for mock and infected organoids at 12 and 24 hpi. C. Dot plot of the top marker genes for each cell type for (left) colon and (right) ileum-derived organoids. The dot size represents the percentage of cells expressing the gene; the color represents the average expression across the cell type. D. Bar plot displaying the proportion of each cell type in mock and infected organoids (12 and 24 hpi).
Interestingly when the researchers treated the cells with UV-inactivated SARS-CoV-2-interferon (IFN), an interferon-stimulated gene (ISG) production did not occur. This implies that active virus replication is required to induce an IFN-mediated response.
Importantly the proinflammatory response occurs by upregulation of the NFκB and TNF pathways - this is clearly observed in the infected intestinal cells.
The bystander cells do not activate the proinflammatory response, as observed in other similar studies.
A moderated upregulation of IFN expression in infected cells in the current model is observed, while a strong ISG upregulation is observed in bystander cells.
The study team also compared the immune response generated by organoids derived from different GI tract sections: 1) ileum organoids were more immunoresponsive than colon organoids, 2) a uniform up-regulation of proinflammatory genes across, and 3) ileum organoids, particularly bystander cells, produced more ISGs comparatively.
.jpg)
Schematic of SARS-CoV-2 infection of human intestinal epithelial cells. SARS CoV-2 infects a subpopulation of enterocytes. Upon infection, enterocytes mount a pro inflammatory response characterized by the upregulation of NFκB and TNF. Bystander cells respond to secreted IFN and upregulate the expression of ISGs. SARS-CoV-2 infection induces the downregulation of ACE2 expression and interferes with IFN-mediated signaling in infected cells.
These observations agree with the previously known fact that different sections of the GI tract respond differently to microbial challenges.
The research findings reveal that the production of ISGs is mostly restricted to the bystander while the production of IFN is detected mostly in infected cells. The latter also fails to produce ISGs and become refractory to IFN stimulus.
Importantly this implies that the SARS-CoV-2 has developed proviral mechanisms to shutdown IFN-mediated signaling and the subsequent production of ISGs in infected cells.
When SARS-CoV-2-infected intestinal cells were treated with IFN, only bystander cells upregulated ISG while infected cells did not. This absence of ISG induction in infected cells suggests that SARS-CoV-2 has developed mechanisms to shutdown IFN-mediated signaling and the subsequent production of ISGs. Preventing IFN-mediated signaling in infected cells would provide a replication advantage to SARS-CoV-2 as secreted IFN will not be able to act in an autocrine manner to induce ISGs which will curtail virus replication and de novo virus production. Although the SARS-CoV-2 viral protein responsible for blocking the IFN-mediated signaling is yet to be identified in our system, a recent report suggests that ORF6 could block IFN-mediated signaling by interfering with STAT1 nuclear translocation. https://www.nature.com/articles/s41467-020-17665-9
Thus this reveals a replication advantage for de novo virus production by curtailing the signaling of antiviral pathways.
This is a significant study where the cellular and the molecular players are identified in the SARS-CoV-2 infection of the human intestinal model.
This study takes us forward in understanding the complete pathogenesis during the interference of SARS-CoV-2 in the intestines of COVID-19 patients.
For more on COVID-19 Immunology, keep on logging to Thailand Medical News
.jpg)

.jpg)