COVID-19 LATEST: European Research Indicates That Precision Medicine Approach Using Blood Transcriptomes Analysis Could Improve COVID-19 Treatments
Source: COVID-19 Latest Jul 12, 2020 5 years, 5 months, 2 weeks, 4 days, 17 hours, 26 minutes ago
COVID-19 Latest: More than 55 European researchers from 13 research centers across Germany, Netherlands and Greece provide the first evidence for the use of whole blood transcriptomics to distinguish COVID-19 disease from other infections, and also to monitor and potentially predict disease outcomes. Such as approach helps further dissect molecular phenotypes and stratify COVID-19 patients, as well as support drug target prediction for specific subgroups of patients.
.jpg) It also indicates that the SARS-CoV-2 coronavirus mode of attacking individuals is very complex yet unique and every patient should be handled from a more personalized approach to study how the virus is affecting them specifically and tailoring a specific treatment protocol, this is where precision medicine come into play.
It also indicates that the SARS-CoV-2 coronavirus mode of attacking individuals is very complex yet unique and every patient should be handled from a more personalized approach to study how the virus is affecting them specifically and tailoring a specific treatment protocol, this is where precision medicine come into play.
Blood transcriptomic profiling studies measure the abundance of blood RNA transcripts on a genome-wide scale. Changes in blood transcript abundance reflect pathogenic processes and inform about the status of the immune system. An RNA sequence mirrors the sequence of the DNA from which it was transcribed. Consequently, by analyzing the entire collection of RNA sequences in a cell (the transcriptome) researchers can determine when and where each gene is turned on or off in the cells and tissues of an organism
The study findings are published on a preprint server but are currently being peer-reviewed.
https://www.medrxiv.org/content/10.1101/2020.07.07.20148395v1
The coronavirus pandemic, which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is still increasing at an alarming pace and extent. Clinical presentations can range from asymptomatic and mild respiratory tract infections to severe cases with acute respiratory distress syndrome, subsequent respiratory failure, and death.
Anomalous global spread and varying symptoms and manifestations urgently demands a more comprehensive molecular understanding of the disease pathophysiology. And while an effective vaccine candidate is still elusive, therapeutic management of COVID-19 patients is pivotal in mitigating the clinical burden, as well as decreasing mortality rates.
The reports on a dysregulated immune system and cytokine storm in severe cases of the disease calls for an improved understanding of the changes found within the immune system to dissect all the parameters accompanying such heterogeneous disease presentation.
The research team aimed to elucidate whether whole blood transcriptomes may be used to determine immune cellular characteristics in COVID-19 patients, reveal heterogeneous molecular phenotypes, commonalities, and differences to other inflammatory conditions, as well as to inform drug repurposing research.
The study team profiled whole blood transcriptomes of 39 COVID-19 patients and 10 control donors between March 13 and March 30, 2020, which in turn enabled a data-driven stratification based on molecular phenotypes.
Interestingly, hierarchical clustering of the most variable genes in the obtained dataset hinted towards further heterogeneity and granularity among patients beyond the current clinical distinction into
mild and severe patients, revealing five patient subgroups.
In a 'reverse transcriptome approach,' the researchers utilized the specific changes found in the COVID-19-related transcriptional modules as the bait and explored the inverse correlation in thousands of drug-based transcriptome signatures to predict prospective drug candidates.
Professor Dr Thomas Ulas from the department of Genomics and Immunoregulation at University of Bonn and the corresponding author told Thailand Medical News, "Further, it became evident that, apart from common therapeutic avenues to address the immune dysregulation in COVID-19 patients, there are patient groups that may benefit from treatments targeting more precisely their immune phenotype and this phenotyping could be used for the enrichment of patient groups in clinical trials.”
The detailed comparison of COVID-19 blood transcriptomes with a collection of over 2,600 samples (derived from 11 different viral infections, inflammatory diseases, and independent control samples) revealed highly characteristic transcriptome signatures for COVID-19.
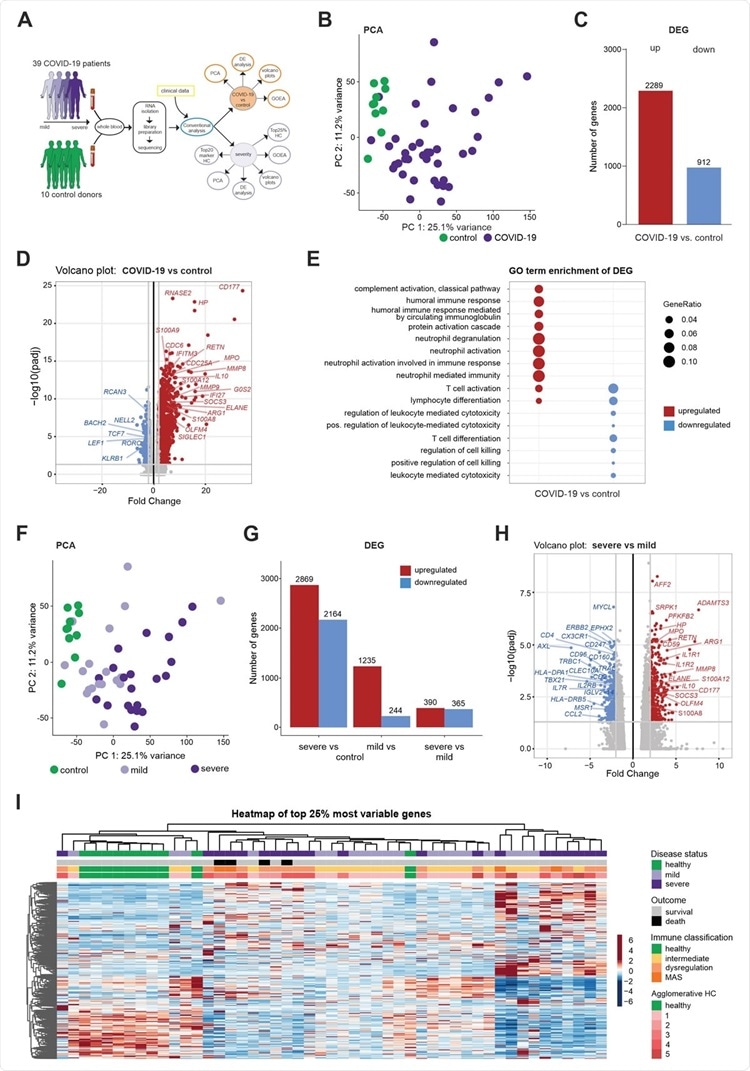 Whole blood transcriptomes reveal a diversity of COVID-19 patients not explained by disease severity (A) Schematic workflow for the analysis of whole blood transcriptome data. (B) PCA plot depicting the relationship of all samples based on dynamic gene expression of all genes comparing COVID-19 and control samples. (C) Number of significantly upregulated (red) and downregulated (blue) genes (FC>|2|, FDR-adj. p-value <0.05) comparing COVID-19 and control samples. (D) Volcano plot depicting fold changes (FC) and FDR-adjusted p-values comparing COVID-19 and control samples. Differentially expressed up- (red) and downregulated genes (blue) are shown and selected genes are highlighted. (E) A plot of top 10 most enriched GO terms for significantly up- and downregulated genes, showing the ratio of significantly regulated genes within enriched GO terms (GeneRatio). (F) PCA plot depicting the relationship of all samples based on dynamic gene expression of all genes comparing mild and severe COVID-19 as well as control samples. (G) Number of significantly upregulated (red) and downregulated (blue) genes (FC >|2|, FDR-adj. p-value < 0.05) comparing mild and severe COVID-19 as well as control samples. (H) Volcano plot depicting fold changes and FDR-adjusted p-values comparing mild and severe COVID-19 as well as control samples. Differentially expressed up- (red) and downregulated genes (blue) are shown and selected genes are highlighted. (I) Hierarchical clustering map of 25% most variable genes between control patients, COVID-19 mild or severe patients, with additional annotation of disease outcome, hierarchical agglomerative clustering of clinical parameters COVID-19, and the groups defined by agglomerative clustering.
Whole blood transcriptomes reveal a diversity of COVID-19 patients not explained by disease severity (A) Schematic workflow for the analysis of whole blood transcriptome data. (B) PCA plot depicting the relationship of all samples based on dynamic gene expression of all genes comparing COVID-19 and control samples. (C) Number of significantly upregulated (red) and downregulated (blue) genes (FC>|2|, FDR-adj. p-value <0.05) comparing COVID-19 and control samples. (D) Volcano plot depicting fold changes (FC) and FDR-adjusted p-values comparing COVID-19 and control samples. Differentially expressed up- (red) and downregulated genes (blue) are shown and selected genes are highlighted. (E) A plot of top 10 most enriched GO terms for significantly up- and downregulated genes, showing the ratio of significantly regulated genes within enriched GO terms (GeneRatio). (F) PCA plot depicting the relationship of all samples based on dynamic gene expression of all genes comparing mild and severe COVID-19 as well as control samples. (G) Number of significantly upregulated (red) and downregulated (blue) genes (FC >|2|, FDR-adj. p-value < 0.05) comparing mild and severe COVID-19 as well as control samples. (H) Volcano plot depicting fold changes and FDR-adjusted p-values comparing mild and severe COVID-19 as well as control samples. Differentially expressed up- (red) and downregulated genes (blue) are shown and selected genes are highlighted. (I) Hierarchical clustering map of 25% most variable genes between control patients, COVID-19 mild or severe patients, with additional annotation of disease outcome, hierarchical agglomerative clustering of clinical parameters COVID-19, and the groups defined by agglomerative clustering.
Furthermore, neutrophil activation-associated signatures were strikingly enriched in patients with severe disease, which was confirmed in whole blood transcriptomes from an independent second cohort and granulocyte samples from a third cohort of COVID-19 patients.
Significantly, there were considerable transcriptional changes with the loss of T cell activation and simultaneous gain of a unique combination of neutrophil activation signals in the blood of severe COVID-19 patients. This observation was not merely due to changes in cell numbers since isolated neutrophils demonstrated identical transcriptional modifications.
Simply by providing crucial insights into the patient structure in COVID-19 and pursuing comparative analysis, this study revealed the first evidence for the unique changes prompted by this disease within the host in comparison to other infections.
The study opens the door for the use of reverse transcriptomics not only in cancer (where it repeatedly proves its value) but also to identify drugs that may target the immune pathophysiology in infections caused by SARS-CoV-2.
Implications for treatment and outcome prediction
This detailed and extensive research endeavor supports the notion that whole blood transcriptomics is not only a way forward for elucidating the systemic immune response in COVID-19 patients, but can also be used to predict novel therapeutic targets which involving distinct pathophysiological mechanisms.
Dr Ulas added, "Most interestingly, we identified drug candidates that might be beneficial for all COVID-19 patients, but also candidates that might only be suitable for a subgroup of patients. And by comparing the transcriptional modules identified in whole blood of COVID-19 patients, we identified unique differences to other viral and bacterial infections, for which similar data were available.”
The study suggests that blood transcriptomes might indeed be very useful for diagnostic purposes or for outcome prediction in different models, larger clinical cohorts, and vaccine trials in the near future.This type of data might be utilized as a starting point for a large-scale assemblage of molecular data during current and future treatment trials for COVID-19 patients based on whole blood transcriptomes.
The study also has implications for the use of Pharmacogenomics in developing personalized treatment protocols.
For more
COVID-19 Latest news, keep on logging to Thailand Medical News.
.jpg)
