COVID-19 Mutations: Study That Tracks Mutation Patterns Of SARS-Cov-2 Warns Of Highly Possible Changes On Nucleocapsid And 3a Viroporin Proteins
Source: COVID-19 Mutations Aug 26, 2020 5 years, 5 months, 1 week, 1 day, 56 minutes ago
COVID-19 Mutations: The SARS-CoV-2 since its debut in China in December 2019, is constantly evolving and mutating to ensure its survival. Unlike most viruses that upon series of mutations gradually losses its virulence and infectivity potential and sometimes simply ‘dies out, the SARS-CoV-2 coronavirus is evolving in a direction that seems to make it more stable and one of that it reasons it that it is equipped with a gene (NSP13 helicase protein) that helps it proof-read the replication of its proteins to make sure only ‘viable’; mutations are retained.
.jpg)
The Bioinformatics Department of the University of Illinois has been studying the mutation patterns of the SARS-CoV-2 coronavirus and keeping track of all the various mutations.]
In a new study, the researchers warn of regions of the virus proteome that is becoming more variable through time, which they say may give us an indication of what to expect next with COVID-19.
Significantly, the study team found increasing mutations in the nucleocapsid protein, which packages the virus's RNA after entering a host cell, and the 3a viroporin protein, which creates pores in host cells to facilitate viral release, replication, and virulence.
The study team says these are regions to watch, because increasing non-random variability in these proteins suggests the virus is actively seeking ways to improve its spread.
Dr Gustavo Caetano-Anolles, Study Senior Author and Professor of Bioinformatics Department, University of Illinois explained to Thailand Medical News that these two proteins interfere with how our bodies combat the virus.
They are the main blockers of the beta-interferon pathway that make up our antiviral defenses. Their mutation could explain the uncontrolled immune responses responsible for so many COVID-19 deaths.
The researchers warn that that possible mutations which seems most likely and not in a positive manner, could bode a very serious and catastrophic outcome, constant attention and monitoring needs to be addressed to watch out for this.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2020.07.31.231472v1
The study shows that the virus is honing the tactics that may make it more successful and more stable.
Graduate students in a spring-semester Bioinformatics and Systems Biology class at University of Illinois tracked the mutation rate in the virus's proteome - the collection of proteins encoded by genetic material, through time, starting with the first SARS-CoV-2 genome published in January and ending more than 15,300 genomes later in May.
The study team found some regions still actively spinning off new mutations, indicating continuing adaptation to the host environment. But the mutation rate in other regions showed signs of slowing, coalescing around single versions of key proteins.
Professor Caetano-Anolles added, “That is bad news. The virus is changing and changing, but it is keeping the things that are most useful or interesting for itself."
;
Interestingly, however, the stabilization of certain proteins could be good news for the treatment of COVID-19.
First author Dr Tre Tomaszewski, a doctoral student in the School of Information Sciences at Illinois said,"In vaccine development, for example, you need to know what the antibodies are attaching to. New mutations could change everything, including the way proteins are constructed, their shape. An antibody target could go from the surface of a protein to being folded inside of it, and you can't get to it anymore. Knowing which proteins and structures are sticking around will provide important insights for vaccines and other therapies."
The study team documented a general slowdown in the virus's mutation rate starting in April, after an initial period of rapid change. This included stabilization within the spike protein, those pokey appendages that give coronaviruses their crowned appearance.
Significantly, within the spike, the researchers found that an amino acid at site 614 was replaced with another (aspartic acid to glycine), a mutation that took over the entire virus population during March and April.
Dr Tomaszewski adds, "The spike was a completely different protein at the very beginning than it is now. You can barely find that initial version now,"
The S or spike protein, which is organized into two main domains, is responsible for attaching to human cells and helping inject the virus's genetic material, RNA, inside to be replicated. The 614 mutation breaks an important bond between distinct domains and protein subunits in the spike.
Dr Caetano-Anolles said, "For some reason, this must help the virus increase its spread and infectivity in entering the host. Or else the mutation wouldn't be kept."
Studies have indicated that the 614 mutation was associated with increased viral loads and higher infectivity in a previous study, with no effect on disease severity.
However, in another study, the mutation was linked with higher case fatality rates.
https://onlinelibrary.wiley.com/doi/full/10.1111/ijcp.13525
This was a hot topic recently in South-East Asia where so called ‘experts’ from Singapore disputed Malaysia’s claims that the mutated version called D614 was more deadly despite not being able to furnish any supporting studies or evidence.
Dr Tomaszewski says although its role in virulence needs confirmation, the mutation clearly mediates entry into host cells and therefore is critical for understanding virus transmission and spread.
Importantly, sites within two other notable proteins also became more stable starting in April, including the NSP12 polymerase protein, which duplicates RNA, and the NSP13 helicase protein, which proofreads the duplicated RNA strands.
Dr Caetano-Anolles added, "All three mutations seem to be coordinated with each other. They are in different molecules, but they are following the same evolutionary process."
However the researchers are more concerned about the new pathways of entropic expansion of mutations involving intrinsically disordered regions of the SARS-CoV-2 proteome and interactions with replicating genomes and endoplasmic membranes that are needed for virus assembly and release from infected cells.
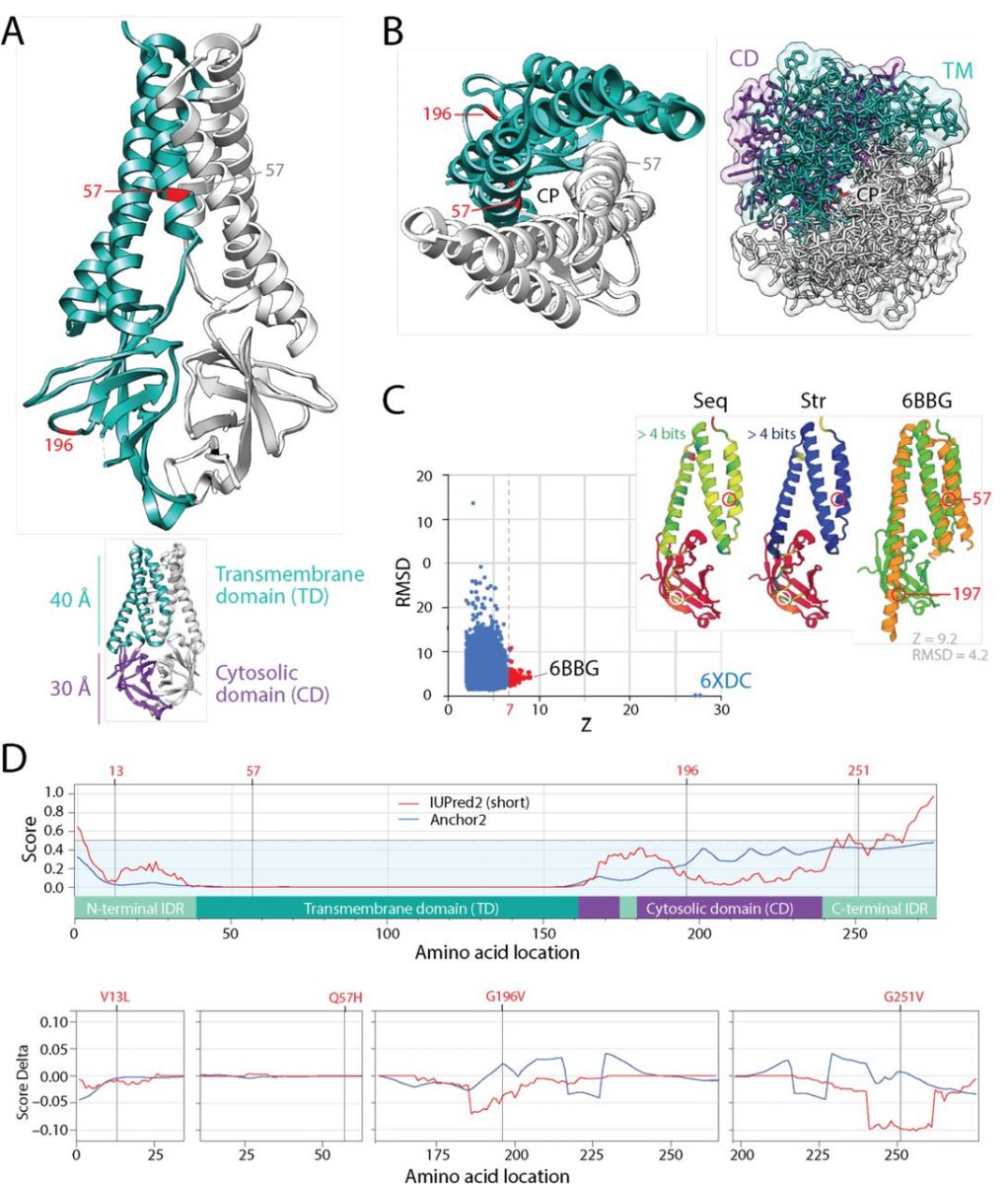 The mutational diversification of SARS-CoV-2 viroporin encoded by ORF3a. A. The structure of the protein 3a molecule (PDB entry 6XDC) has two domains, a N-terminal transmembrane domain (TD) and a C-terminal cytosolic domain (CM). Mutation Q57H is located in the first of the three transmembrane helices at the major hydrophilic constriction of the pore important for channel activity. Mutation G197V forms part of a loop at the surface of the CD. Terminal amino acids 1-38 and 239-275 and a 175-180 in CD could not be modeled because they were weakly resolved. They hold mutations 13 and 251. B. View from the lumen side of the channel pore (P) in ribbon and atom stick representation. Note that the pore is only 1 Å wide. C. A DALI structural neighborhood analysis (10,088 structural neighbors) returned significant hits to small fragments (Z ≤ 9.2; RMSD ≥ 1.3) that formed a single cluster in the RMSD versus Z-score plot. Structural alignment of the 92 hits with Z ≥ 7 (red dots) revealed that all hits matched the TD structures and were well conserved at structure (Str) but less at sequence (Seq) levels. The best structural match to the TD was the Orai protein channel (PDB entry 6BBG) responsible for Ca2+ influx pathways in metazoan cells and involved in immune responses and cancer. D. The mapping of intrinsic disorder (UIPred2, red line) and gain-loss of binding energy (Anchor2, blue line) along the sequence confirmed the significant intrinsic disorder (scores ≥ 0.5) of the C-terminal linker. A comparison of the different mutants and reference viral strain with a delta score revealed that mutations G196V and G251V decreased disorder
The mutational diversification of SARS-CoV-2 viroporin encoded by ORF3a. A. The structure of the protein 3a molecule (PDB entry 6XDC) has two domains, a N-terminal transmembrane domain (TD) and a C-terminal cytosolic domain (CM). Mutation Q57H is located in the first of the three transmembrane helices at the major hydrophilic constriction of the pore important for channel activity. Mutation G197V forms part of a loop at the surface of the CD. Terminal amino acids 1-38 and 239-275 and a 175-180 in CD could not be modeled because they were weakly resolved. They hold mutations 13 and 251. B. View from the lumen side of the channel pore (P) in ribbon and atom stick representation. Note that the pore is only 1 Å wide. C. A DALI structural neighborhood analysis (10,088 structural neighbors) returned significant hits to small fragments (Z ≤ 9.2; RMSD ≥ 1.3) that formed a single cluster in the RMSD versus Z-score plot. Structural alignment of the 92 hits with Z ≥ 7 (red dots) revealed that all hits matched the TD structures and were well conserved at structure (Str) but less at sequence (Seq) levels. The best structural match to the TD was the Orai protein channel (PDB entry 6BBG) responsible for Ca2+ influx pathways in metazoan cells and involved in immune responses and cancer. D. The mapping of intrinsic disorder (UIPred2, red line) and gain-loss of binding energy (Anchor2, blue line) along the sequence confirmed the significant intrinsic disorder (scores ≥ 0.5) of the C-terminal linker. A comparison of the different mutants and reference viral strain with a delta score revealed that mutations G196V and G251V decreased disorder
These pathways involve intrinsically disordered regions of the N-protein, a structural protein that forms complexes with genomic RNA, interacts with the Mprotein during virus assembly, enhances the efficiency of virus transcription and assembly, and help overcome the host innate immune response.
These pathways also involve viroporins. Viroporins of enveloped viruses such as SARS-CoV-2 insert into membranes to break chemoelectrical barriers by channeling ions across membranes and dissipating membrane potential, a property that stimulates budding and resembles that of depolarization-dependent exocytosis.
The close homology of SARS-CoV-2 protein 3a to the Orai proteins suggests membrane potential dissipation involves store-operated channeling mechanisms that control Ca2+ cellular levels. These new mutational pathways may be responsible for new symptomatic manifestations of the COVID-19 disease. For example, asymptomatic SARS-CoV-2 infection has been reported to account for ~50% of total coronavirus cases and the majority of the patients with mild infection symptoms can ‘recover’ by themselves. But not much is known how the subsequent virus reservoirs in the human host or the viral proteins affect the human host and the resulting long term health complications.
Also the COVID-19 disease mechanism suggests that the severe symptoms of COVID-19 involve the uncontrolled immune response of the host. Indeed, mutational pathways involve virus molecules that can subvert the immune response, specifically the interferon response. For example, the N-protein, protein 3a and accessory protein 6 are the three b-interferon antagonists operating in coronavirus disease. Mutations in two of these molecules are high entropy in our mutational set. In addition, the SARS-CoV-2 new virus tends to be more rapidly spreading and less lethal (in terms of immediate reactions but again we do not know about the long term health complications) than SARS-CoV and MERS-CoV, a fact that demands explanation.
Dr Tomaszewski commented, “As predicted by the director of Centers for Disease Control and Prevention of the USA, this virus is going to be with us. I am hopeful that we will get through this first wave and have some time to prepare for the second wave. We hope the exploration of mutational pathways can anticipate moving targets for speedy therapeutics and vaccine development as we prepare for the next wave of the pandemic.”
He added that along with thousands of other researchers sequencing, uploading, and curating genome samples through the GISAID Initiative, the team at University of Illinois will continue to keep track of this virus.
For more on
COVID-19 Mutations, keep on logging to Thailand Medical News.
.jpg)
