COVID-19 News: Coronavirus Seems To Not Be Able To Tolerate Indians! Study Claims Numerous Mutations In India With Virus Weakening And A Possible Extinction
Source: COVID-19 News Aug 20, 2020 4 years, 8 months, 6 days, 11 hours, 7 minutes ago
COVID-19 News: It is becoming obvious that the SARS-CoV-2 coronavirus is changing as it spreads to different countries and regions. This is due spontaneous adaptive mutations, shaped by the different selection pressures exerted by varying environmental conditions and hosts.
.jpg)
A new study by Indian researchers from the JIS Institute of Advanced Studies and Research-Kolkata and the School of Biotechnology, Amrita Vishwa Vidyapeetham, Kollam-Kerala, shows that the SARS-CoV-2 coronavirus is rapidly mutating in India with the emergence of various strains, all showing weakening attributes, leading experts to speculate that the novel coronavirus might eventually become extinct in India.
The study findings that have yet to have been peer-reviewed is published on a preprint server.
https://www.biorxiv.org/content/10.1101/2020.08.18.255570v1
The study was initially focused on finding and interpreting the association between the evolution of the SARS-CoV-2 and the conditions in the Indian environment and hosts.
The study team looked at how the viral spike glycoprotein showed mutation-derived differences since this is the key to viral binding, fusion, and cell entry leading to host cell infection.
Significantly, this is the key pathogenetic feature that differentiates it from earlier pathogenic coronaviruses SARS-CoV and MERS-CoV. It also carries the two most important genetic features of the virus.
“By examining the architecture of the spike glycoprotein and its mechanics, one can reveal the susceptibility of the virus and can foresee the facts helping in the discovery of the SARS-CoV-2 antidote." one of the co-researchers. commented.
The study team utilized the genomic sequences of 630 Indian isolates retrieved from the GISAID database. Tracing these mutations showed that the spike protein variants followed two pathways starting from two ancestral strains, namely, Wuhan-Hu-1/2019 and its D614G variant.
Although the Wuhan-Hu-1/2019 strain gave rise to the D614G variant, it also evolved to 20 other variants, of which one further led to three others.
On the other hand, the other ancestral strain D614G gave rise to 47 variants, of which 9 led to further variants.
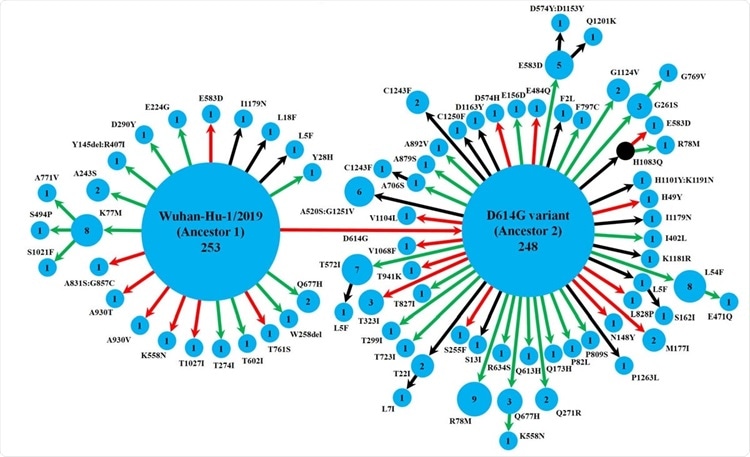 Schematic representation of the diversity of spike protein variants circulating in India, using maximum likelihood-based phylogeny reconstruction. Each node represents a specific spike protein variant, while the node-size and the number inside depict the frequency of that variant. The red or green color of each arrow indicates the higher or lower stability index respectively of the S-R complex for each variant than the major ancestral variant it emerged from (either Ancestor 1 or Ancestor 2). The black arrows lead to the variants for which the docking scores could not be determined either because of the presence of at least one variation outside the available template regio
n for docking(18, 42), or due to non-existing isolate in the lone hypothetical node with H1083Q mutation denoted by black color. This black node signifies a variant with no available isolate in our dataset, while it gives rise to two derived variants, H1083Q:R78M and H1083Q:E583D, for which representative isolates were available
Schematic representation of the diversity of spike protein variants circulating in India, using maximum likelihood-based phylogeny reconstruction. Each node represents a specific spike protein variant, while the node-size and the number inside depict the frequency of that variant. The red or green color of each arrow indicates the higher or lower stability index respectively of the S-R complex for each variant than the major ancestral variant it emerged from (either Ancestor 1 or Ancestor 2). The black arrows lead to the variants for which the docking scores could not be determined either because of the presence of at least one variation outside the available template regio
n for docking(18, 42), or due to non-existing isolate in the lone hypothetical node with H1083Q mutation denoted by black color. This black node signifies a variant with no available isolate in our dataset, while it gives rise to two derived variants, H1083Q:R78M and H1083Q:E583D, for which representative isolates were available
Both ancestral strains are predominant, but there were 16 detectable spike protein variants found in multiple isolates, with different levels of spike-receptor stability. Strikingly, several mutations occurred independently at the same position across eight variants, which were not linked phylogenetically.
Significantly, more than 50 percent of these variants were found exclusively only in India, but of these, over two-thirds had lower spike-receptor stability than the ancestral strains.
The study team says that this is expected in a region where a new virus that is rapidly mutating tries to adapt to a host. However both positive and detrimental mutations may occur without reversion in the absence of any genetic recombination mechanism.
Importantly, the implication is that this rapid buildup of mutations in the more recent strains leads to a loss of fitness compared to their ancestors, despite the constant increase in the rate of mutations.
The study team invoked Muller’s ratchet, the hypothesis that in asexual reproduction, the irreversible accumulation of harmful mutations causes a species to die out gradually.
Being an RNA virus, the SARS-CoV-2 coronavirus inherently has a high mutation rate. Understanding how this is occurring at the molecular level could help understand how it causes disease and how it can be curbed.
Interestingly, the most significant spike protein diversity was in Maharashtra, Odisha, West Bengal, and Gujarat, but this is limited by the fact that many other states had very few isolates.
The study team said that this diversity could be due to positive selection pressures enabling the protein, which is a prime immune target, to evade host immunity and allow entry into the cell. This could affect spike-receptor binding. The team found 41 and 22 mutations in the S1 and S2 subunits of the spike, respectively.
The detailed study of mean spike-receptor complex stability in each state, based on the individual stability indices of the variants in circulation in that state, was then performed using the docking score of the S1 subunit with the receptor.
By limiting it to only those with 50 or more sequenced isolates, they looked at this index in Maharashtra, Delhi, Gujarat, and Telangana, which account for 70% of the total isolates, and cover a wide range of diversity.
The team explained that the mean stability of the spike-receptor complex across these variants was strongly and exponentially correlated with fatality rates (deceased: recovered cases) across the Indian subcontinent.
Significantly, the fatality rate dropped by seven times between April 11 and June 28, 2020.
It was also observed that Telangana and Delhi had similar average stabilities at a 7% fatality rate, while Maharashtra and Gujarat showed exponentially lower stabilities, with 8% and 9% fatality rates, respectively.
The study team says that this suggests that the S-R complex stability is a potential marker to assess the severity of the disease.
The team also suggest that further study is necessary at the population level to validate this experimentally, especially since the actual parameter (the docking score) used has not been directly correlated with the fatality rate so far.
.jpg) Heat map distribution across four Indian states with >50 sequenced isolates based on average stability index. The average stability index for a particular state denotes the averaged value of docking scores / HADDOCK scores of S-R complexes for all circulating variants. The values of average stability index and fatality rate in Indian states are plotted to fit an exponential function (R2=0.96).
Heat map distribution across four Indian states with >50 sequenced isolates based on average stability index. The average stability index for a particular state denotes the averaged value of docking scores / HADDOCK scores of S-R complexes for all circulating variants. The values of average stability index and fatality rate in Indian states are plotted to fit an exponential function (R2=0.96).
The study team put forth the theory that this loss of stability may be responsible for the lower national fatality rate. In India, after the initial phase in which the fatality steadily went up for four weeks, to peak at 38% on April 11, 2020, it declined sharply until it reached 5% on June 28, 2020, the date at which the researchers last obtained their data. However, the mutation rate was increasing during this period from March to May 2020.
The researchers suggest, “Could Muller’s ratchet be a player in shaping SARS-CoV-2 evolutionary dynamics in India?”
Implying that the virus could be gradually extinguishing itself under the accumulating weight of deleterious mutations, without the washout process of natural selection being able to compensate because of the rapid rate of mutations. This causes harmful mutations to be fixed in the viral population, and this buildup could lead to a “
mutational meltdown.”
Interestingly the short period of the study precludes a definitive answer, but the trend may be seen, according to the researchers.
Importantly, this could also help evolve a better therapeutic approach, facilitating the meltdown process by inducing more harmful mutations. This will require large-scale genomic studies to answer the question, and thereby helping to improve public health surveillance and prevention programs.
The study team concluded, “While we propose the potential of SR complex stability to track disease severity, we also urge an immediate need to explore if SARS CoV 2 is approaching mutational meltdown in India.”
These rapid virus mutations which making the virus weak might be beneficial for India as the country has a large population with very limited healthcare facilities and resources, these mutations could indirectly save lives and prevent more people in India from having to go through hardships as a result of the COVID-19 crisis.
UPDATE: This study since 2nd of October has not been peer- reviewed positively and has never made it to a journal plus emerging statistics in India is showing that there is no possibility that the virus is weakening nor will become extinct. Read:
https://www.thailandmedical.news/news/breaking-india-news-study-shows-new-mutational-strains-and-d614g-mutation-emerging-in-central-india,-covid-19-crisis-in-india-expected-to-worsen
For more
COVID-19 News, keep on logging to Thailand Medical News.
Help! Please help support this website by kindly making a donation to sustain this website and also all in all our initiatives to propel further research: https://www.thailandmedical.news/p/sponsorship
All donations of value US$50 to US$100 will also receive complimentary therapeutic teas that can be useful in this COVID-19 era. https://www.thailandmedical.news/news/new-therapeutic-teas-
.jpg)

.jpg)