COVID-19 News: French Study Shows That Host Proteins G3BP Plays A Key Proviral Role In SARS-Cov-2 Viral Assembly
Nikhil Prasad Fact checked by:Thailand Medical News Team Jan 22, 2024 1 year, 3 months, 4 days, 8 hours, 13 minutes ago
COVID-19 News: The ongoing global battle against the COVID-19 pandemic has spurred intensive research efforts to decipher the molecular intricacies of the SARS-CoV-2 virus. Recent strides in understanding the virus-host interactions have culminated in a groundbreaking study conducted by researchers at Université Paris Cité, CNRS, Inserm, and Institut Cochin in Paris, France. This study covered in this
COVID-19 News report, delves into the late stages of the viral lifecycle, revealing the unexpected and pivotal proviral role of host proteins G3BP1 and G3BP2 in the assembly of SARS-CoV-2 virions.
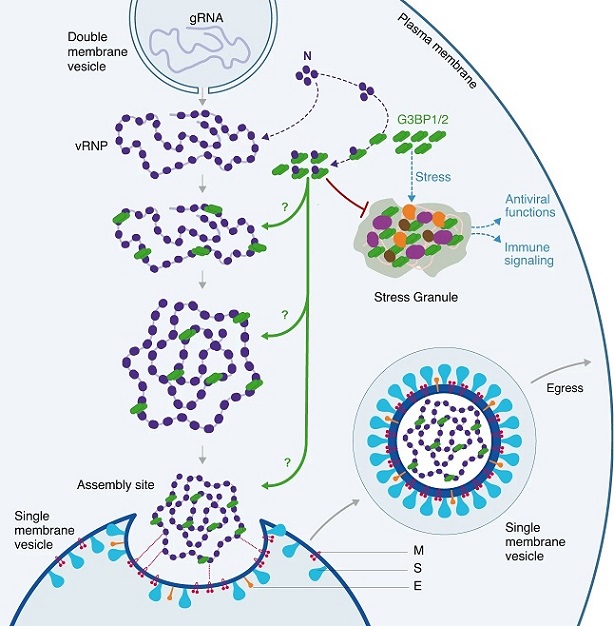 Schematic model of the possible roles of G3BP proteins in the assembly of SARS-CoV-2 virions
Schematic model of the possible roles of G3BP proteins in the assembly of SARS-CoV-2 virions
In a past Thailand
Medical News coverage in 2021, it was found that blocking SARS-C0V-2 peptide motifs protein interactions with G3BP proteins could inhibit infections.
https://www.thailandmedical.news/news/breaking-scientists-discover-blocking-protein-interactions-between-human-g3bp1-2-proteins-and-sars-cov-2-xfg-peptide-motif-inhibits-infection
Upon stress and in the absence of SARS-CoV-2 infection, G3BP proteins can trigger phase separation of ribonucleoprotein complexes (RNPs) and induce stress granule (SG) assembly. SG are sites of RNA storage. Upon viral infections, they can also sequester viral factors, control viral translation, stimulate immune signaling or prevent apoptosis. Upon SARS-CoV-2 infection, N interacts with G3BP1/2 and viral genomic RNA (gRNA), preventing SG formation. The study findings indicate that G3BP1/2 also favor the assembly of new infectious virions possibly by: 1-participating to the viral RNP (vRNP) stabilization and/or reorganization, 2-inducing phase separation and vRNP compaction or 3-favoring the recruitment of the vRNP to the assembly site and its packaging.
The Imperative to Understand SARS-CoV-2 Replication
As the causative agent of the COVID-19 pandemic, SARS-CoV-2 has posed unprecedented challenges to global public health. While rapid vaccine development has provided a claimed robust defense against severe pathologies, there remains a need for prophylactic treatments. Furthermore, emerging evidence suggests that the virus can persist in certain tissues, potentially contributing to long-COVID. To mitigate these challenges and curb the emergence of more contagious strains, a deeper understanding of the molecular mechanisms underlying SARS-CoV-2 replication is imperative.
Molecular Landscape of SARS-CoV-2 Replication
Upon entry into host cells, the SARS-CoV-2 genomic RNA (gRNA) is translated in the cytoplasm, giving rise to polyproteins that undergo cleavage to genera
te non-structural proteins (nsps). These nsps assemble, recruiting host cell proteins to form a replication and transcription complex within double-membrane vesicles (DMVs). The RNA-dependent RNA polymerase utilizes gRNA as a template, synthesizing both progeny genomic RNA and subgenomic RNAs. The newly synthesized RNAs are exported through transmembrane pores of DMVs, subsequently used for the translation of viral proteins. The viral N protein plays a pivotal role in genome packaging, forming ribonucleoproteins (RNPs) along the genomic RNA in a bead-on-a-string conformation. This unique feature is crucial for packaging the unusually large SARS-CoV-2 gRNA into virions.
Viral RNPs are then recruited by the M protein and assemble with other structural proteins at modified cellular membranes derived from the endoplasmic reticulum (ER), the Golgi, and the reticulum-Golgi intermediate compartment (ERGIC), along with surrounding single membrane vesicles (SMVs). Finally, the newly formed virions are released into SMVs and exported to the plasma membrane.
Host-Virus Interactions: A Comprehensive Approach
Given the reliance of SARS-CoV-2 on cellular machinery for virion production, understanding host-virus interactions is crucial. While previous large-scale screens focused on early stages of viral replication, there was a notable gap in investigating the late stages, particularly gRNA packaging, particle assembly, and egress. To address this, the researchers conducted a proteomic analysis of SARS-CoV-2 particles produced from A549-ACE2 and Calu-3 cells, isolating virions through ultracentrifugation on sucrose cushion and ACE-2 affinity capture.
A Comprehensive Dataset: Insights into Virion Production
The proteomic analysis resulted in the identification of 356 putative host factors associated with SARS-CoV-2 virions. These factors underwent stringent criteria, including their involvement in molecular and cellular pathways linked to viral infections, interactions with other virion-associated factors, and their presence in other viral particles. Notably, 92 host factors were identified as embedded within virions, providing a rich resource for understanding the molecular environment of virion production.
Stress Granule Proteins: Unexpected Participants
Among the identified host factors, a subset of stress granule proteins caught the researchers' attention. Stress granules are cytoplasmic membraneless structures that form in response to cellular stress, including viral infections. G3BP1 and G3BP2, known as major nucleators of stress granules, were specifically enriched in SARS-CoV-2 virions. This unexpected finding prompted further exploration into the role of G3BP1 and G3BP2 in the viral assembly process.
G3BP Proteins: Dual Functionality in Viral Replication
Contrary to the conventional understanding of stress granules as antiviral factors, the study revealed a proviral role for G3BP1 and G3BP2 in SARS-CoV-2 replication. These proteins actively contributed to the later stages of the viral lifecycle, such as gRNA packaging, virion assembly, or the egress of newly infectious viral particles.
The researchers demonstrated that G3BP1 and G3BP2 downregulation resulted in a decrease in SARS-CoV-2 replication. Interestingly, disrupting the interaction between the viral N protein and G3BP1 altered stress granule assembly, indicating the virus's ability to modulate host cell responses for its benefit.
Mechanistic Insights and Future Implications
To unravel the impact of G3BP1 and G3BP2 on SARS-CoV-2 virion production, the researchers utilized two pulmonary cell models, A549-ACE2 and Calu-3 cells. Simultaneously knocking down both G3BP1 and G3BP2 revealed a consistent decrease in the production of infectious virions. This led to the conclusion that these proteins play a crucial role in gRNA packaging, virion assembly, or the egress of newly infectious viral particles.
The study proposes a model wherein G3BP1 and G3BP2 interact with the viral N protein and/or gRNA, promoting virion assembly and/or accumulation in cytoplasmic vesicles. This dual functionality of G3BP proteins, both as antiviral factors through stress granule assembly and as pro-viral factors in virion assembly, suggests a complex interplay between the virus and host cell machinery.
Implications for Therapeutics: Targeting G3BP Proteins
The unexpected proviral role of G3BP1 and G3BP2 in SARS-CoV-2 replication opens new avenues for potential therapeutic interventions. Targeting the interaction between these host proteins and viral components could be a promising strategy to dampen viral replication. The study suggests the development of drugs that prevent N/G3BP interaction, providing a potential dual benefit by favoring stress granule formation and hindering SARS-CoV-2 virion assembly.
Conclusion
In conclusion, the groundbreaking study illuminates the crucial role of host proteins G3BP1 and G3BP2 in the late stages of SARS-CoV-2 replication. By unraveling the intricate mechanisms of viral assembly, this research not only enhances our understanding of the virus-host interplay but also provides valuable insights into potential therapeutic targets for controlling the spread of the virus. As the global scientific community continues its collaborative efforts against the COVID-19 pandemic, delving into the molecular landscape of host-virus interactions remains paramount for developing effective treatments and preventive measures.
The study findings were published in the peer reviewed journal: Nature Communications.
https://www.nature.com/articles/s41467-024-44958-0
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
Read Also:
https://www.thailandmedical.news/news/covid-19-news-scientists-discover-yet-an-additional-mechanism-by-which-sars-cov-2-disarms-host-immune-responses,-nsp-inhibits-stress-granule-formation
https://www.thailandmedical.news/news/study-uncovers-that-sars-cov-2-n-proteins-are-able-to-enter-stress-granules-and-cause-amyloid-aggregation,-leading-to-various-neurodegenerative-issues
https://www.thailandmedical.news/news/covid-19-news-sars-cov-2-nsp3-hijacks-fragile-x-mental-retardation-proteins-for-efficient-infection-while-modulating-stress-granule-function
