COVID-19 News: International Study Finds That SARS-CoV-2 NSP14 Governs Mutational Instability And Plays Key Role In The Emergence Of New Variants!
Nikhil Prasad Fact checked by:Thailand Medical News Team Jan 13, 2024 1 year, 3 months, 5 days, 14 hours, 53 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the rapid spread of the SARS-CoV-2 virus, has brought about an urgent need for in-depth research to understand the mutational dynamics driving the emergence of new variants. Among the multitude of proteins encoded by the virus, the Nonstructural protein 14 (NSP14) has emerged as a key player in the evolution of SARS-CoV-2 variants. This
COVID-19 News report delves into the findings of an international study, shedding light on the multifaceted role of NSP14, its influence on mutational instability, and its potential as a therapeutic target.
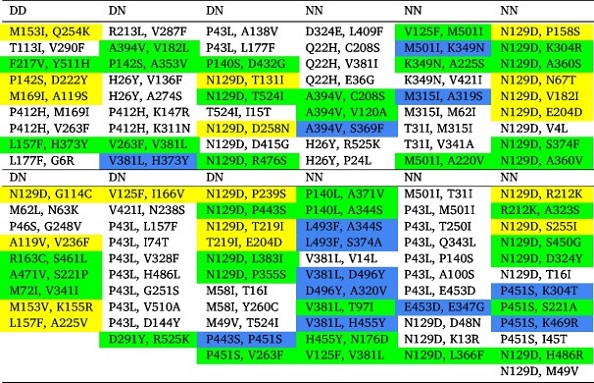 Three different classes of co-occurring mutations in SARS-CoV-2 NSP14 variants.Yellow, blue, and green marked co-occurring mutations belong to the ExoN, N7-MTase, and both (one was from ExoN & another was from N7-MTase) domains, respectively.
NSP14 Mutations Drive SARS-CoV-2 Variant Evolution
Three different classes of co-occurring mutations in SARS-CoV-2 NSP14 variants.Yellow, blue, and green marked co-occurring mutations belong to the ExoN, N7-MTase, and both (one was from ExoN & another was from N7-MTase) domains, respectively.
NSP14 Mutations Drive SARS-CoV-2 Variant Evolution
At the heart of SARS-CoV-2's adaptability lies the multifunctional NSP14 enzyme, equipped with both exonuclease and messenger RNA (mRNA) capping capabilities. The study reveals that NSP14 significantly influences the replication fidelity of SARS-CoV-2. Single and co-occurring mutations within NSP14 play a pivotal role in driving mutational instability, contributing to the emergence of diverse SARS-CoV-2 variants. These mutations impact the virus's ability to replicate with accuracy, giving rise to the genetic diversity observed in different viral strains.
Genetic Diversity in NSP14 Variants
A comprehensive analysis of 3953 NSP14 variants categorizes them into three distinct clusters, underscoring the substantial genetic diversity inherent within the virus. This research goes beyond mere categorization, identifying 120 co-mutations, 68 unique mutations, and 160 conserved residues across NSP14 homologs. Such insights illuminate the intricate web of phylogenetic patterns, pathogenicity factors, and residue interactions, providing a deeper understanding of the virus's evolutionary landscape.
Functional Significance of NSP14 Domains
Understanding the genetic makeup of SARS-CoV-2 is crucial for deciphering its behavior and evolution. The virus, a member of the Coronaviridae family, belongs to the betacoronavirus group and is characterized by its positive-sense, single-stranded RNA genome. The study emphasizes the significant sequence similarities between SARS-CoV-2 and its predecessors, MERS-CoV (2013) and SARS-CoV (2001). It highlights the high transmissibility of SARS-CoV-2, leading to its devastating global impact.
NSP14 Structure and Function
Delving into the intricacies of NSP14, the study provides an in-depth look at the bifunctional role it plays within SARS-CoV-2. Comprising approximately 527 amino acids, NSP14 contains two distinct domains - the ExoN domain at the N-terminal and the N7-MTase domain at the C-terminal. The ExoN domain is particularly noteworthy for its proofreading activity, ensuring high-fidelity replication of the virus. This critical function reduc
es error rates and maintains genomic integrity, essential for the virus's survival.
The N7-MTase domain, on the other hand, plays a crucial role in mRNA capping, allowing the virus to evade host immune responses. Mutations in key residues of both domains have far-reaching consequences, affecting viral replication, immune evasion, and the overall evolution of the pathogen. The study highlights the conservation of certain residues across different coronaviruses, indicating shared mechanisms in RNA substrate recognition and suggesting potential targets for drug development.
The study places a spotlight on the critical roles of the two distinct domains of NSP14 - the proofreading ExoN domain and the N7-MTase domain responsible for RNA capping. Mutations in key residues, particularly within the ExoN domain, can significantly impact viral replication. This detailed exploration of the structural and functional aspects of NSP14's domains unveils potential drug targets for therapeutic interventions. The ExoN domain, with its proofreading function, emerges as a focal point for potential drug development strategies.
Balance of Deleterious and Neutral Mutations
An intricate analysis of NSP14 sequences reveals a delicate balance between 548 neutral mutations and 414 deleterious mutations. The interplay between these mutations is crucial for viral fitness and adaptation. Deleterious mutations, which result in a loss of gene function or impaired protein production, play a role in down-regulating the host immune response. This balance becomes a dynamic force shaping the survivability and evolutionary trajectory of SARS-CoV-2.
Previous Investigations and Limitations
While previous investigations into NSP14 have yielded invaluable insights into its roles in replication fidelity, immune evasion, and viral evolution, the article acknowledges certain limitations. Earlier studies often provided a broad overview of NSP14 mutations without detailed geographic analysis, hindering a comprehensive understanding of the enzyme's behavior across diverse locations. The study aims to address these limitations and contribute a nuanced and geographically informed understanding of NSP14 mutations.
Contributions of the Study
This COVID-19 News report outlines the significant contributions of the present study, offering a detailed analysis of NSP14 mutations. It identifies a total of 962 single mutations in SARS-CoV-2 NSP14, categorizing 414 as deleterious and 548 as neutral mutations. The study also uncovers 160 invariant residues through sequence homology analysis, providing potential drug design targets.
Additionally, the investigation into the polarity of invariant residues sheds light on their potential stabilizing roles, contributing to a more comprehensive understanding of coronavirus protein interactions.
The study further explores 120 co-occurring mutations across different geographic locations, categorizing them based on their impact. The identification of structural and functional clusters among SARS-CoV-2 NSP14 sequences adds another layer of complexity to our understanding, aiding in deciphering the evolutionary implications and trends between different variants.
Implications for Therapeutic Development
While acknowledging the limitations associated with the lack of comprehensive functional validation, the study identifies potential drug targets within NSP14, especially within the ExoN domain. The findings underscore the urgent need to explore NSP14-ExoN as a potential inhibition target for novel antiviral strategies, particularly in the context of SARS-CoV-2 resistance to existing antivirals like nucleoside analogs. The multifunctional role of NSP14 emerges as a focal point for understanding the diverse outcomes and long-term symptoms associated with COVID-19.
As the study concludes, it points out the necessity of further research to validate the impact of co-mutations on viral fitness and adaptation. The challenges posed by SARS-CoV-2's resistance to existing antivirals, particularly nucleoside analogs, necessitate a deeper exploration of targeting NSP14-ExoN as a novel inhibition target. Understanding the multifunctional role of NSP14, especially its ExoN activity, not only offers insights into the virus's replication fidelity but also presents a promising avenue for the development of novel antiviral strategies.
Moreover, the findings suggests that the study's insights may be crucial in future investigations into the primary sequelae observed in other coronaviruses (SARS, MERS), including respiratory, musculoskeletal, and neuropsychiatric issues. The multifunctional enzyme NSP14 within SARS-CoV-2 could play a significant role in influencing these outcomes. Investigating the involvement of NSP14 in these aspects may shed light on the diverse outcomes and pathophysiological mechanisms associated with COVID-19.
In summary, the international study on SARS-CoV-2 NSP14 provides a thorough examination of the enzyme's role in viral evolution, mutational dynamics, and potential therapeutic targeting. The findings contribute significantly to our understanding of SARS-CoV-2 and offer promising avenues for further research and therapeutic development in the ongoing battle against the COVID-19 pandemic.
The study findings were published in the peer reviewed journal: Computers in Biology and Medicine.
https://www.sciencedirect.com/science/article/pii/S0010482523013641
The study involved scientists from the following institutions: Pingla Thana Mahavidyalaya,Bengal-India, Indian Institute of Science Education and IISER Berhampur Transit campus, Odisha-India, Indian Statistical Institute,West Bengal-India, Pondicherry University-India, University of the Witwatersrand, Johannesburg,-South Africa, Woxsen University, Telangana-India, King Abdulaziz University, Jeddah-Saudi Arabia, Genetic Engineering and Biotechnology Research Institute, Alexandria,-Egypt, PanTherapeutics-Switzerland, Institute of Integrative Omics and Applied Biotechnology (IIOAB), Nonakuri-India, Federal University of Minas Gerais-Brazil, State University of Southwest of Bahia (UESB)-Brazil-University of Lincoln-UK, Yarmouk University-Jordan, International University of Sarajevo-Bosnia and Herzegovina, Federal University of Rio de Janeiro (UFRJ)-Brazil, Universidad Católica de Valencia San Vicente Mártir-Spain, University of South Florida, Tampa-USA and Ras Al Khaimah Medical and Health Sciences University-United Arab Emirates.
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
