COVID-19 News: Mexican Study Shows That Variants Of Host Genes IFNAR1 And IFNAR2 Play Role In The Immune Response Against SARS-CoV-2
Nikhil Prasad Fact checked by:Thailand Medical News Team Nov 07, 2023 2 years, 2 months, 3 weeks, 3 days, 1 hour, 1 minute ago
COVID-19 News: The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has underscored the critical role of genetics in influencing susceptibility to the virus and the severity of the resulting disease. Among the myriad of genetic factors at play, single-nucleotide variants (SNVs) in specific host genes have emerged as key determinants of COVID-19 susceptibility and severity. In particular, genes encoding the type I interferon receptor (IFNAR), IFNAR1, and IFNAR2, have come under scrutiny for their impact on the immune response to the virus. This
COVID-19 News report explores the findings of a systematic review conducted by researchers at the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas in Mexico, shedding light on the significance of SNVs in these genes and their potential implications for COVID-19 management.
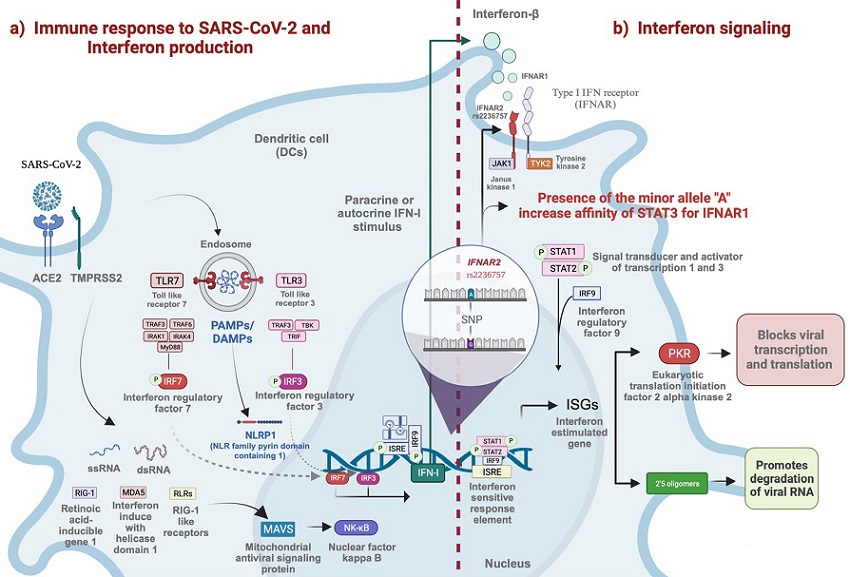 Entry of SARS-CoV-2 into the cell via ACE2 and TMPRSS2. The endosome is formed by double-membrane vesicles where the coronavirus replicates. Viral RNA (ssRNA, dsRNA) is recognized by TLR3, TLR7, RIG-I, and MDA5. These receptors activate TRIF and MyD88, downstream TLRs, and MAVS downstream RIG-I and MDA5, starting in the interferon production pathway. When IRF3, IRF7, or NFkB are activated, they translocate to the nucleus and trigger the transcription of immunogens like inflammatory cytokines and IFN. The dotted line represents the transition to IFN signaling, beginning with IFN-I binding IFNAR to initiate JAK/STAT signaling. In silico assays propose that the A allele of rs2236757 in IFNAR2 increases the affinity of STAT3 to IFNAR1.
Entry of SARS-CoV-2 into the cell via ACE2 and TMPRSS2. The endosome is formed by double-membrane vesicles where the coronavirus replicates. Viral RNA (ssRNA, dsRNA) is recognized by TLR3, TLR7, RIG-I, and MDA5. These receptors activate TRIF and MyD88, downstream TLRs, and MAVS downstream RIG-I and MDA5, starting in the interferon production pathway. When IRF3, IRF7, or NFkB are activated, they translocate to the nucleus and trigger the transcription of immunogens like inflammatory cytokines and IFN. The dotted line represents the transition to IFN signaling, beginning with IFN-I binding IFNAR to initiate JAK/STAT signaling. In silico assays propose that the A allele of rs2236757 in IFNAR2 increases the affinity of STAT3 to IFNAR1.
The IFNAR1 and IFNAR2 genes encode subunits of the type I interferon receptor, a vital component of the body's antiviral defense system. These subunits, IFNAR1 and IFNAR2, play distinct roles in the receptor complex. IFNAR2, characterized by a high affinity for interferons, is a membrane-bound protein that, when activated, initiates signaling cascades involving Janus kinases (JAK2) and signal transducers and activators of transcription (STATs). The activation of STATs leads to their translocation to the nucleus and subsequent transcription of interferon-stimulated genes (ISGs), which are critical for antiviral responses.
Type I interferons, such as IFN-α and IFN-β, trigger the production of various IFN-stimulated gene products with antiviral, antiproliferative, and immunomodulatory functions. One such product is the RNA-activated protein kinase (PKR), which becomes activated in the presence of viral double-stranded RNA, ultimately reducing viral protein synthesis. IFNAR signaling also leads to the production of 2′ - 5′ oligoadenylate (2′ - 5′A), which activates RNase and, in turn, degrades viral RNA.
Genetic variants in IFNAR1 and IFNAR2 can disrupt the delicate balance of this signaling pathway, leading to decreased protein abundance or impaired internalization in response to interferon binding. For instance, homozygosity for a nonsense variant in IFNAR2 was reported in two siblings who developed severe encephalitis after receiving a live-attenuated measles, mumps, and rubella (MMR) immunization. Other common single-nucleotide variants (SNVs) in IFNA
R1 and IFNAR2, such as rs201609461, rs1601861199, and rs1601861196, can result in truncated or modified proteins with impaired functionality, potentially impacting the immune response to viral infections.
The emergence of SARS-CoV-2, the virus responsible for COVID-19, has brought the significance of these genetic variants to the forefront. As the virus enters the airway and infects alveolar epithelial cells in the lungs, the host's immune system is activated, and the IFN signaling pathway plays a crucial role. While SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) receptor for entry into host cells, the virus is detected by dendritic cells through Toll-like receptor 7 (TLR7) in the endosome, and cytosolic RNA sensors such as RIG-I and MDA-5 recognize viral double-stranded RNA during replication.
Upon infection with SARS-CoV-2, signaling cascades are initiated, resulting in the production of type I and type III interferons, which contribute to the antiviral immune response. However, SARS-CoV-2 has developed various mechanisms to evade the interferon pathway, such as the repression of type I interferon (IFN-β) activation by viral proteins like ORF6, ORF8, and the nucleocapsid protein. These viral proteins interfere with the signaling pathways, ultimately inhibiting the host's immune response and promoting viral replication.
The clinical manifestations of COVID-19 vary widely, with symptoms ranging from mild to severe, including pneumonia, acute respiratory distress syndrome, organ damage, and even death. At present, there is no curative treatment for COVID-19, but certain strategies have shown promise in specific patient populations. In the early stages of COVID-19, a triple combination therapy involving interferon beta-1b, lopinavir/ritonavir, and ribavirin has demonstrated efficacy in reducing symptoms, accelerating viral clearance, and shortening hospital stays in mild to moderate cases.
Nevertheless, several factors, such as age, gender, and comorbidities, affect the clinical course and outcomes of COVID-19. Genetic variations, particularly single-nucleotide variants (SNVs), play a significant role in determining an individual's susceptibility to infection and the severity of the disease. Population-based studies, including genome-wide association studies (GWAS) and candidate gene studies, have been pivotal in identifying SNVs associated with COVID-19 susceptibility and severity.
The systematic review conducted by researchers in Mexico aimed to elucidate the role of SNVs in IFNAR1 and IFNAR2 in COVID-19 susceptibility and severity. Through an exhaustive search of the literature, they identified 11 relevant studies that included case reports of rare SNVs (defined by minor allele frequency < 1%) and GWAS. The review aimed to answer a crucial research question: Do the risk variants in SNVs of IFNAR1 or IFNAR2 in patients with severe COVID-19 affect the antiviral immune response or lead to poor outcomes compared to non-carriers of these risk variants?
The study provided several compelling insights into the impact of genetic variants in IFNAR1 and IFNAR2. It revealed that certain rare variants in IFNAR2, such as rs72550721, lead to a loss of function in IFNAR2, making individuals more susceptible to severe COVID-19.
Furthermore, the presence of specific SNVs in IFNAR1, such as rs9975538 and rs13050728, was associated with increased expression of IFNAR2, potentially conferring protection by enhancing the antiviral activity. Notably, these findings suggest that the response to IFN therapy may also be influenced by genetic variants, as seen with rs9984273 in IFNAR2, where individuals with specific alleles showed differential responses to interferon treatment.
The researchers highlighted the importance of identifying high-risk groups that could benefit from IFN-α/IFN-β therapy. Such therapy has shown promise in inhibiting SARS-CoV-2 replication and modulating the immune response. The trial of a combination therapy including interferon beta-1b, lopinavir/ritonavir, and ribavirin demonstrated its safety and efficacy in patients with mild to moderate COVID-19.
Understanding the genetic variations in IFNAR1 and IFNAR2 is crucial for tailoring treatment strategies, especially in cases where the interferon response is compromised. Furthermore, research in this area underscores the potential of genetic variants to guide therapeutic decisions, ultimately improving patient outcomes.
In conclusion, single-nucleotide variants in the IFNAR1 and IFNAR2 genes can significantly impact the immune response to SARS-CoV-2 infection, potentially exacerbating or attenuating the disease. These genetic variants represent promising targets for therapies designed to limit infection and mitigate the resulting inflammation. As the world continues to grapple with the COVID-19 pandemic, the insights gained from studies like the one conducted in Mexico provide valuable knowledge that may inform more precise and effective approaches to managing this global health crisis.
The study findings were published in the peer reviewed journal: Pathogens.
https://www.mdpi.com/2076-0817/12/11/1320
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
