COVID-19 News: Spanish Study Finds That SARS-CoV-2 Remodels The Human Host Landscape Of Small Noncoding RNAs! Possible Serious Health Complications!
Nikhil Prasad Fact checked by:Thailand Medical News Team Oct 02, 2023 1 year, 6 months, 3 weeks, 3 days, 17 hours, 38 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has had a profound impact on global health, emphasizing the critical need for a deeper understanding of the host response to this viral infection. In a groundbreaking study conducted by the University of Valencia in Spain and the Clinic University Hospital, INCLIVA Biomedical Research Institute-Spain, researchers have delved into how SARS-CoV-2 reshapes the landscape of small non-coding RNAs (sncRNA) using a vast collection of nasopharyngeal swab samples collected at various time points from patients displaying different levels of symptom severity.
 COVID-19 News: Spanish Study Finds That SARS-CoV-2 Remodels The Human Host Landscape Of Small Noncoding RNAs! Possible Serious Health Complications!
COVID-19 News: Spanish Study Finds That SARS-CoV-2 Remodels The Human Host Landscape Of Small Noncoding RNAs! Possible Serious Health Complications!
To date, no studies or
COVID-19 News reports have covered this area of SARS-CoV-2 possibly affecting human host small non-coding RNAs (sncRNA) and this study is the first.
The study utilized high-throughput RNA sequencing to analyze the sncRNA profiles, revealing a global alteration in sncRNA landscape during SARS-CoV-2 infection, characterized by peaks in abundance related to RNA species of 21-23 and 32-33 nucleotides. By employing principal component analysis, the researchers successfully differentiated infected patients based on their sncRNA profiles. Notably, host-derived sncRNAs, including microRNAs (miRNAs), transfer RNA-derived small RNAs (tsRNAs), and small nucleolar RNAs (sdRNAs), exhibited significant differential expression in infected patients compared to control subjects. MiRNA expression, in particular, was predominantly downregulated in response to SARS-CoV-2 infection, particularly in patients with severe symptoms.
The study also identified specific tsRNAs derived from Glu- and Gly-tRNAs as major altered elements upon infection, with 5' tRNA halves being the most abundant species. Additionally, downregulation of C/D-box sdRNAs and altered expression of tyRNAs were observed in infected patients. These findings offer valuable insights into the host sncRNA response to SARS-CoV-2 infection, potentially paving the way for the development of diagnostic and therapeutic strategies in clinical settings.
Understanding the Transcriptomic Response to SARS-CoV-2
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 triggered a global pandemic, leading to a surge in cases of coronavirus disease 2019 (COVID-19). This highly contagious virus primarily targets the respiratory system, causing a range of symptoms, from mild respiratory distress to severe pneumonia and multi-organ failure. While significant efforts have been made to elucidate the pathogenesis and pathophysiology of COVID-19, our understanding of the transcriptomic changes that occur during infection remains limited, particularly in the noncoding RNA (ncRNA) realm.
Noncoding RNAs are a diverse group of RNA molecules that do not code for proteins but play crucial roles in the regulation of gene expression. Small noncoding RNAs (sncRNAs), typically less than 50 nucleotides in length, are a subset of ncRNAs with dive
rse functions in cellular processes. These include microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), transfer RNA-derived small RNAs (tsRNAs), small nucleolar RNAs (sdRNAs), and trimmed forms of miRNAs known as tiny RNAs (tyRNAs). While initially considered byproducts of RNA metabolism, sncRNAs are now recognized as key players in gene regulation.
MiRNAs, for example, are generated from structured RNA precursors and can inhibit gene expression by binding to complementary mRNA targets, leading to translation repression or mRNA degradation. PiRNAs are involved in maintaining genomic integrity and regulating transposable elements. TsRNAs, derived from transfer RNAs, have been associated with stress responses, cancer, and viral infections. SdRNAs guide the cleavage of target RNAs, while tyRNAs are thought to enhance RNA cleavage processes. The intricate roles of these sncRNAs are still being uncovered, making them an area of active research.
Analyzing SARS-CoV-2's Impact on sncRNA Landscape
In this study, researchers sought to comprehensively investigate how SARS-CoV-2 infection remodels the human sncRNA landscape by analyzing clinical samples collected from nasopharyngeal swabs of COVID-19 patients at different stages of infection and with varying degrees of symptom severity. The study focused on two time points: an initial time when the first symptoms appeared (T1, approximately 5-7 days post-infection) and a later time (T2, approximately 19-21 days post-infection).
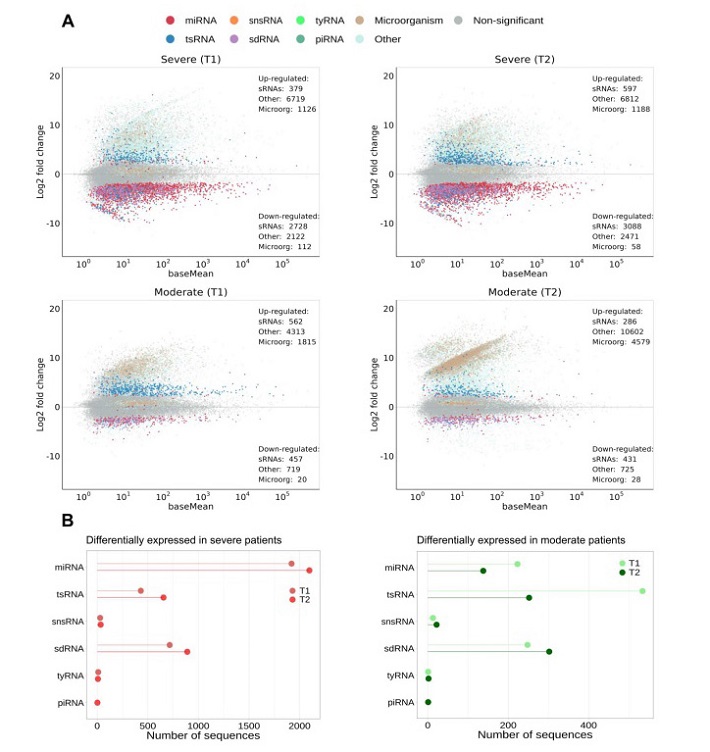
To provide a global view of the sncRNA landscape, RNA was extracted from these samples and subjected to high-throughput sequencing. Subsequent analysis revealed distinct sncRNA profiles in response to SARS-CoV-2 infection, as demonstrated by principal component analysis, which effectively distinguished infected patients from control subjects. The majority of sncRNAs identified in the study were host-derived elements, with a small proportion corresponding to microorganism-derived elements and an even smaller fraction matching the SARS-CoV-2 genome.
Notably, the length distribution of the identified sncRNA reads showed peaks at 21-23 nucleotides, a characteristic length range for miRNAs, and an additional peak at 32-33 nucleotides, suggesting alterations in sncRNA populations during infection.
Differential Expression of sncRNAs in Response to Infection
The researchers then conducted pairwise comparisons between control and infected samples to identify sncRNAs with significant differential expression upon SARS-CoV-2 infection. These differentially expressed sncRNAs were categorized based on their homology to various classes of regulatory sncRNAs, including miRNAs, tsRNAs, piRNAs, tyRNAs, sdRNAs, and snsRNAs.
The results revealed a substantial alteration in sncRNA expression during SARS-CoV-2 infection. Notably, miRNAs, sdRNAs, and tsRNAs showed consistent alterations in all analyzed samples, with miRNAs being the most affected. MiRNA downregulation was particularly pronounced in patients with severe symptoms, both at the early and late stages of infection. In contrast, tsRNAs exhibited upregulation as a general response to infection, with the extent of accumulation varying based on disease severity and time of infection. Specific tsRNAs derived from Glu- and Gly-tRNAs were identified as major elements affected by infection, particularly in severe cases.
Additionally, the study observed changes in the expression of tyRNAs, a relatively unexplored class of sncRNAs. While only a few tyRNAs were differentially expressed, they exhibited both upregulation and downregulation in response to infection, with variations depending on the severity and stage of COVID-19.
Notably, the study identified six miRNA families consistently responding to SARS-CoV-2 infection, including hsa-let-7, hsa-miR-182, hsa-miR-183, hsa-miR-205, hsa-miR-2110, and hsa-miR-16. These findings are significant as they shed light on specific miRNA families that play critical roles in the host's response to the virus, potentially offering avenues for further research and therapeutic development.
Glu- and Gly-tRNA-derived tsRNAs emerged as a major focus of the study, with 5' tRNA halves being the most abundant species upon infection. These tsRNAs may hold promise as potential biomarkers for the presence of the virus and the prediction of disease severity. Furthermore, the observed downregulation of C/D-box sdRNAs and the altered expression of tyRNAs provide additional insights into the complex interplay between sncRNAs and SARS-CoV-2 infection.
Implications and Future Directions
The findings of this Spanish study represent a significant contribution to our understanding of the host response to SARS-CoV-2 infection at the sncRNA level. The comprehensive analysis of sncRNA profiles in patients with varying disease severity and at different stages of infection provides valuable insights that can inform the development of diagnostic and therapeutic strategies.
The downregulation of specific miRNAs, such as hsa-let-7, in response to SARS-CoV-2 infection suggests their potential role in the host's defense mechanisms or susceptibility to infection. Understanding the precise functions of these miRNAs and their targets could lead to novel therapeutic approaches.
In humans and other mammals, hsa-let-7 is involved in the regulation of critical physiological processes, such as organ development, growth, tissue regeneration, metabolism, cancer proliferation and modulation of viral infection. Downregulation of has-let-7 can lead to a whole serious of heath complications including the rise of certain cancers!
The differential expression of tsRNAs, particularly those derived from Glu- and Gly-tRNAs, highlights their potential as biomarkers for viral presence and disease severity prediction. Further research into the functional roles of these tsRNAs in the context of SARS-CoV-2 infection is warranted.
Additionally, the study's discovery of altered sdRNAs and tyRNAs in response to SARS-CoV-2 infection opens up new avenues of investigation into the regulatory roles of these sncRNAs during viral infections. Clarifying the functions of these sncRNAs could provide critical insights into the mechanisms of host-virus interactions.
It is important to note that the disruptions and dysregulation of these various non-coding RNAs (sncRNA) and also miRNAs can lead to serious health complications and issues.
In conclusion, this study underscores the importance of exploring the noncoding RNA landscape in the context of viral infections like COVID-19. As our understanding of sncRNA functions and their roles in host-pathogen interactions continues to grow, we may uncover novel diagnostic tools and therapeutic targets for managing SARS-CoV-2 infection and future viral outbreaks.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.researchsquare.com/article/rs-3375685/v1
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
