COVID-19 News: Study Shows That SARS-CoV-2 ORF3a-Mediated NF-KB Activation Is Not Dependent On TRAF Binding Sequence
Nikhil Prasad Fact checked by:Thailand Medical News Team Nov 09, 2023 2 years, 1 month, 2 weeks, 1 day, 8 hours, 14 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has swept across the globe, infecting millions and leading to countless deaths. As scientists and researchers grapple with understanding the virus and its effects on the human body, a recent study conducted by Case Western Reserve University in Cleveland, USA, in collaboration with the University of Science and Technology of China in Hefei, China, sheds light on a critical aspect of the virus's pathogenic mechanisms. This groundbreaking research delves into the role of SARS-CoV-2's accessory protein, open reading frame 3a (ORF3a), in the activation of the nuclear factor kappa B (NF-κB) pathway. Notably, this study covered in this
COVID-19 News report challenges previous findings related to ORF3a and its interaction with TNF receptor-associated factor (TRAF) proteins, providing new insights into the potential mechanisms driving excessive inflammation in COVID-19.
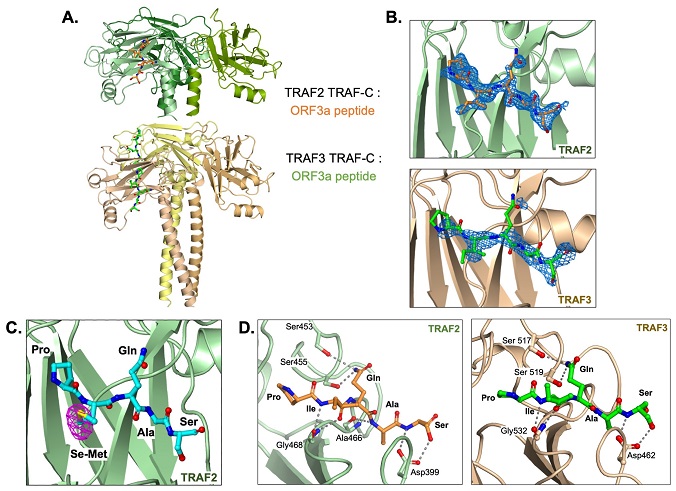 TRAF2 and TRAF3 TRAF-C domains in complex with ORF3a peptide. (A) Trimer structure of TRAF2 TRAF-C (upper) or TRAF3 TRAF-C (lower) in complex with one or two ORF3a peptides, respectively. Five peptide residues, PIQAS, are shown as orange or green ball-and-stick models in the TRAF2 and TRAF3 structures, respectively. (B) Fo-Fc omit map (blue mesh, 2.5 sigma) in the peptide-binding groove of TRAF2 (upper panel) or TRAF3 (lower panel) with PIQAS residues from the ORF3a peptide superimposed. (C) Residues from selenomethionine (Se-Met)-containing ORF3a peptide in complex with the TRAF2 TRAF-C domain. Anomalous difference map (magenta mesh) is shown at 4 sigma. (D) Hydrogen bonds between residues in ORF3a peptide and residues in TRAF2 (left) or TRAF3 (right) TRAF-C domains are shown as gray dotted lines.
SARS-CoV-2 and the Inflammatory Response
TRAF2 and TRAF3 TRAF-C domains in complex with ORF3a peptide. (A) Trimer structure of TRAF2 TRAF-C (upper) or TRAF3 TRAF-C (lower) in complex with one or two ORF3a peptides, respectively. Five peptide residues, PIQAS, are shown as orange or green ball-and-stick models in the TRAF2 and TRAF3 structures, respectively. (B) Fo-Fc omit map (blue mesh, 2.5 sigma) in the peptide-binding groove of TRAF2 (upper panel) or TRAF3 (lower panel) with PIQAS residues from the ORF3a peptide superimposed. (C) Residues from selenomethionine (Se-Met)-containing ORF3a peptide in complex with the TRAF2 TRAF-C domain. Anomalous difference map (magenta mesh) is shown at 4 sigma. (D) Hydrogen bonds between residues in ORF3a peptide and residues in TRAF2 (left) or TRAF3 (right) TRAF-C domains are shown as gray dotted lines.
SARS-CoV-2 and the Inflammatory Response
The COVID-19 pandemic has posed a significant challenge to healthcare systems worldwide. The virus has infected over three-quarters of a billion people, with millions succumbing to the disease or its complications. One of the defining features of severe COVID-19 is the excessive inflammation that can lead to a cytokine storm and severe tissue damage, particularly in the lungs. However, despite extensive research efforts, the pathogenesis of COVID-19 remains not fully understood. While much attention has been devoted to the virus's structural proteins, such as the Spike protein, the SARS-CoV-2 genome encodes sixteen non-structural proteins and nine accessory proteins. These accessory proteins may play vital roles in the development of COVID-19.
The Role of ORF3a in COVID-19 Pathology
Among these accessory proteins, ORF3a has emerged as a potential key player in COVID-19 pathology. Previous studies have shown that the deletion of ORF3a in a mouse model of SARS-CoV-2 infection resulted in a lower mortality rate and reduced viral load in the lungs compared to wild-type virus-infected mice.
Furthermore, ORF3a expression in the brains of mice led to neuroinflammation and neurodegeneration. ORF3a is a transmembrane protein with the capacity to form homodimers and homotetramers. It shares a high
degree of structural similarity with the ORF3a protein in the original SARS-CoV, with both proteins exhibiting primary sequence identity and similar secondary, tertiary, and quaternary structures.
ORF3a's multiple proposed functions include inducing cell death, disrupting cellular membrane structures, impairing autophagy, and potentially forming cation channels. While there have been debates regarding the ion conductance properties of ORF3a, it is clear that it can induce inflammatory responses. One notable mechanism is the activation of the NLRP3 inflammasome, a key component of the body's inflammatory response. Activation of the NLRP3 inflammasome typically involves a priming step that activates NF-κB to upregulate the expression of sensor proteins like NLRP3, pro-caspase-1, and pro-IL-1β.
Subsequently, an assembly step involves the oligomerization of NLRP3 and the recruitment and auto-processing of pro-caspase-1, leading to the cleavage of pro-IL-1β and pro-IL-18. Previous research on SARS-CoV ORF3a has shown that it can activate NF-κB during the priming step, and this activation relies on a specific N-terminal sequence known as PIQAS, which conforms to TNF receptor-associated factor (TRAF) binding consensus sequences.
TRAF proteins are essential components of several NF-κB signaling pathways, serving as adaptor proteins and E3 ubiquitin ligases. These proteins interact with receptor intracellular domains and other molecules through their C-terminal TRAF-C domains. Given the conservation of the TRAF-binding sequence (PIQAS) between SARS-CoV and SARS-CoV-2 ORF3a, it was hypothesized that SARS-CoV-2 ORF3a might also bind to TRAFs, thus inducing NF-κB activation through a similar mechanism.
Investigating the Interaction Between SARS-CoV-2 ORF3a and TRAF Proteins
To explore this hypothesis, the researchers conducted a series of experiments. They solved the crystal structures of the TRAF-C domains from TRAF2 and TRAF3 in complex with a SARS-CoV-2 ORF3a peptide containing the TRAF-binding sequence. The results of this analysis confirmed that the TRAF-binding sequence in ORF3a mediated its interaction with the TRAF-C domains, albeit with low affinity. Using fluorescence polarization assays, they determined that the interaction's affinity ranged from 150-200 μM, which is similar to other TRAF-binding peptides.
To investigate whether SARS-CoV-2 ORF3a could activate NF-κB, the researchers employed dual-luciferase experiments. The findings indicated that SARS-CoV-2 ORF3a indeed activated NF-κB when compared to control conditions. However, a critical discovery was that mutating the TRAF-binding sequence did not significantly impact ORF3a-induced NF-κB activation. This observation challenged previous research on SARS-CoV ORF3a, which relied on its TRAF-binding sequence (PLQAS) for NF-κB activation.
Implications and Future Research
The research presented here reveals intriguing insights into the mechanisms by which SARS-CoV-2 ORF3a induces NF-κB activation. While previous studies on SARS-CoV ORF3a had emphasized the importance of its TRAF-binding sequence, it appears that SARS-CoV-2 ORF3a may activate NF-κB through alternative mechanisms. This distinction suggests that the two related viruses may have evolved different strategies for modulating the host's immune response.
The role of ORF3a in COVID-19 is complex, as it is implicated in various signaling pathways, including apoptosis, autophagy, and inflammasome activation.
Additionally, it has been shown to disrupt cellular membrane structures, leading to membrane reorganization. This multifunctional capacity is not unique to SARS-CoV-2 ORF3a but is shared with other coronavirus accessory proteins, such as ORF3 from porcine epidemic diarrhea virus (PEDV). These proteins impact various aspects of cellular physiology and are likely to exert their pathological effects in a cell type- or tissue-specific manner.
Further research is essential to unravel the precise roles and functions of accessory proteins like ORF3a and ORF3 in the context of COVID-19. Understanding these roles could have significant implications for the development of therapeutics and interventions to combat coronavirus infections effectively.
In conclusion, this study conducted by Case Western Reserve University and the University of Science and Technology of China has shed new light on the role of SARS-CoV-2 ORF3a in the activation of NF-κB, a key pathway in the host's inflammatory response. While previous research on related coronaviruses emphasized the importance of the TRAF-binding sequence in ORF3a, this study indicates that SARS-CoV-2 ORF3a may activate NF-κB through alternative mechanisms. These findings open up new avenues for further research into the multifaceted functions of coronavirus accessory proteins and also into possible therapeutics to deal with COVID-19 infections.
The study findings were published in the peer reviewed journal: Viruses.
https://www.mdpi.com/1999-4915/15/11/2229
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
