COVID-19 News: Study Uncovers How SARS-CoV-2 Nucleocapsid Protein Manipulates Innate Immunity Pathways Via Inhibiting MAVS Ubiquitination
Nikhil Prasad Fact checked by:Thailand Medical News Team Dec 12, 2023 1 year, 4 months, 1 week, 6 days, 13 hours, 4 minutes ago
COVID-19 News: The COVID-19 pandemic has thrust the world into an unprecedented global health crisis, prompting intensive research to unravel the intricate molecular mechanisms employed by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to subvert host defenses. Severe cases of COVID-19 are characterized by a compromised interferon (IFN) response, allowing unchecked viral replication and heightened inflammation. While the SARS-CoV-2 nucleocapsid (N) protein has been implicated in impeding innate immunity, the exact mechanism remains elusive. This
COVID-19 News report delves into a groundbreaking study conducted by researchers from Hubei University of Medicine, Capital Medical University, and The First Affiliated Hospital of Jinan University in China, shedding light on the interaction between the SARS-CoV-2 N protein and UBC9, unveiling its role in inhibiting MAVS ubiquitination by enhancing its SUMOylation.
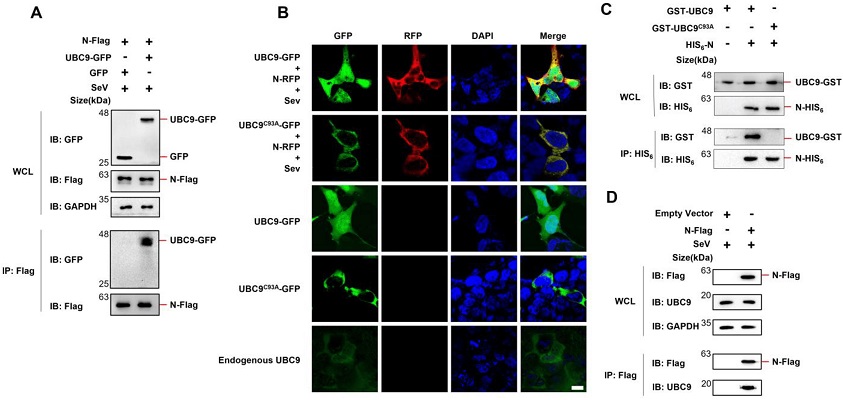 SARS-COV-2 N protein interacts with UBC9. (A) pCDNA3.1 (3XFlag Tag) with N protein-coding genes and pEGFP-N1 (GFP Tag) with UBC9-coding genes were co-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pDsRED-mono-N1 (RFP Tag) with SARS-COV-2 N protein-coding genes and pEGFP-N1 (GFP Tag) with UBC9 or UBC9C93A coding genes were expressed or co-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cells were stained with DAPI before confocal microscopy imaging. Endogenous UBC9 in HEK293T cells was also detected by FITC-conjugated Goat Anti-Mouse IgG(H + L), which recognizes anti-UBC9 mouse antibodies. Scale bars: 10 μm. (C) pET28a (HIS6 tag, Kan+) with SARS-COV-2 N protein coding genes and pGEX-6P-1 (GST tag, Amp+) with UBC9 or UBC9C93A coding genes were co-expressed in BL21 (DE3) E. coli. cell lysates were subjected to Co-IP after IPTG (1 mM) induction for 6 h. (D) pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein-coding genes was over-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cell lysates were collected for Co-IP assays.
Host Defense Responses and Impaired IFN-I Response in COVID-19
SARS-COV-2 N protein interacts with UBC9. (A) pCDNA3.1 (3XFlag Tag) with N protein-coding genes and pEGFP-N1 (GFP Tag) with UBC9-coding genes were co-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pDsRED-mono-N1 (RFP Tag) with SARS-COV-2 N protein-coding genes and pEGFP-N1 (GFP Tag) with UBC9 or UBC9C93A coding genes were expressed or co-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cells were stained with DAPI before confocal microscopy imaging. Endogenous UBC9 in HEK293T cells was also detected by FITC-conjugated Goat Anti-Mouse IgG(H + L), which recognizes anti-UBC9 mouse antibodies. Scale bars: 10 μm. (C) pET28a (HIS6 tag, Kan+) with SARS-COV-2 N protein coding genes and pGEX-6P-1 (GST tag, Amp+) with UBC9 or UBC9C93A coding genes were co-expressed in BL21 (DE3) E. coli. cell lysates were subjected to Co-IP after IPTG (1 mM) induction for 6 h. (D) pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein-coding genes was over-expressed in HEK293T cells. After a 24-h lipofectamine transfection followed by 8 h of SeV stimulation, cell lysates were collected for Co-IP assays.
Host Defense Responses and Impaired IFN-I Response in COVID-19
Upon infection with RNA viruses like SARS-CoV-2, retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) recognize pathogen-associated molecular patterns (PAMPs). This recognition triggers a signaling cascade that culminates in the production of interferons (IFN-β and IFN-α) and sets the stage for antiviral defenses. However, severe COVID-19 patients exhibit a distinct phenotype marked by a severe impairment of the type I interferon (IFN I) response, resulting in a persistent viral load and exacerbated inflammation. The SARS-CoV-2 N protein has been previously implicated in disrupting IFN-β signaling by interacting with RIG-I, hindering the host's innate immune response.
The N Protein's Novel Role: Modulating MAVS SUMOylation
Recent research indicates that the N protein of SARS-CoV-2 goes beyond its previously identified role and interacts wi
th the human SUMO-conjugating enzyme UBC9. This interaction, explored in detail in the study, amplifies the molecular interplay between UBC9 and mitochondrial antiviral signaling protein (MAVS). Specifically, the N protein enhances MAVS SUMOylation, a crucial protein modification process.
Results and Implications
The study unfolds a series of compelling results, providing a comprehensive understanding of the molecular mechanisms orchestrated by the SARS-CoV-2 N protein:
-
Interaction with UBC9: The study confirms a direct interaction between the N protein and the human SUMO-conjugating E2 enzyme UBC9.
-Enhanced UBC9-MAVS Interaction: The N protein, particularly its N-terminal domain (NTD), intensifies the molecular interaction between UBC9 and MAVS.
-Inhibition of MAVS Ubiquitination: The heightened interaction between MAVS and UBC9 inhibits MAVS ubiquitination by promoting its SUMOylation.
-Crucial Role in Regulation: The interaction between the N protein and UBC9 emerges as a pivotal regulator of MAVS SUMOylation and ubiquitination during virus infection.
-Critical Role of UBC9: The UBC9 enzyme plays a critical role in the impaired IFN I response caused by the N protein-MAVS interaction during virus infection.
Discussion: Unraveling the SARS-CoV-2-Nucleocapsid Protein Nexus
The study's findings underscore the complexity of the interplay between SARS-CoV-2 and the host's innate immune response. By elucidating the intricate mechanisms through which the N protein modulates MAVS SUMOylation and ubiquitination, the research paves the way for targeted therapeutic strategies against severe COVID-19.
The ongoing global battle against SARS-CoV-2 necessitates a holistic understanding of the virus-host interactions. The initial steps involve the recognition of viral RNA duplexes by key players like RIG-I, initiating a cascade that leads to the production of interferons. However, severe cases of COVID-19 reveal a compromised IFN response, prompting investigations into the specific viral proteins responsible for disrupting this crucial defense mechanism.
The study's revelation of the interaction between the N protein and UBC9 unveils a novel dimension of SARS-CoV-2's evasion tactics. The enhanced molecular interplay between UBC9 and MAVS, orchestrated by the N protein, inhibits MAVS ubiquitination by promoting SUMOylation. Consequently, this impedes the phosphorylation events crucial for IFN-β signaling, disrupting the host's innate immune response.
Conclusion: Illuminating the Path Forward
In conclusion, the study findings provide a crucial piece of the puzzle in understanding SARS-CoV-2's manipulation of the host's innate immune response. The N protein's role in modulating MAVS SUMOylation and ubiquitination sheds light on the intricate mechanisms employed by the virus to subvert host defenses.
These findings not only contribute to the ongoing efforts to decipher SARS-CoV-2's pathogenesis but also hold promise for the development of targeted therapeutic strategies against severe COVID-19. As the global scientific community continues its relentless pursuit of solutions to the COVID-19 pandemic, each revelation brings us closer to understanding the virus's intricacies and devising effective countermeasures.
The study findings were published in the peer reviewed journal: Viruses.
https://www.mdpi.com/1999-4915/15/12/2304
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
