COVID-19 News: UK Study Finds That SARS-CoV-2 NSP12 Associates With TRiC And The P323L Substitution Acts As A Host Adaption
Nikhil Prasad Fact checked by:Thailand Medical News Team Nov 08, 2023 1 year, 5 months, 1 week, 3 days, 9 hours, 38 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has taken the world by storm, leading to a global health and economic crisis. The virus has undergone numerous genetic changes since its emergence in late 2019, resulting in variants with different properties, including increased transmissibility and the ability to partially evade natural or vaccine-acquired immunity. Among these genetic variations, two significant mutations have garnered particular attention: the D614G substitution in the viral spike protein and the P323L substitution in the viral RNA-dependent RNA polymerase, known as NSP12. These two mutations are at the core of the dominant global landscape of SARS-CoV-2.
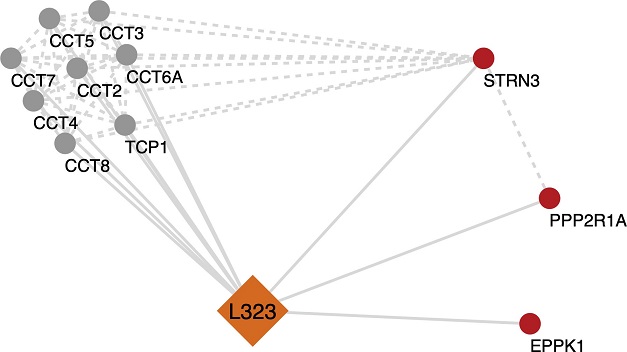 Analysis of selected significant cellular proteins that potentially interact with NSP12L323. Potential protein-protein interaction networks of NSP12L323 were derived using Cytoscape from the mass spectrometry interactome data. Multiple proteins were found to be associated with NSP12L323. In this simplified figure, the highly enriched chaperonin-containing TCP1 (TRiC/CCT complex) is involved in the folding and assembly of proteins associated with the NSP12L323 interactome. A few proteins were identified to be enhanced in the NSP12L323 interactome (STRN3 and PP2R1A, which are part of the PP2A phosphatase family)
Analysis of selected significant cellular proteins that potentially interact with NSP12L323. Potential protein-protein interaction networks of NSP12L323 were derived using Cytoscape from the mass spectrometry interactome data. Multiple proteins were found to be associated with NSP12L323. In this simplified figure, the highly enriched chaperonin-containing TCP1 (TRiC/CCT complex) is involved in the folding and assembly of proteins associated with the NSP12L323 interactome. A few proteins were identified to be enhanced in the NSP12L323 interactome (STRN3 and PP2R1A, which are part of the PP2A phosphatase family)
In this
COVID-19 News report, we delve into a groundbreaking study conducted by the University of Liverpool and the University of Bristol in the United Kingdom, which sheds light on the interaction between NSP12 and host cell proteins, uncovering a potential host adaptation mechanism that influences virus evolution. While much attention has been focused on the spike protein mutation, NSP12's role in the virus's biology is equally pivotal, and the study shows that it forms a complex web of interactions with cellular proteins that contribute to its function and the virus's replication.
The Emergence of SARS-CoV-2 Variants
SARS-CoV-2, which originated in animals and made the jump to humans in the Huanan Seafood Wholesale Market, experienced its first major genetic changes after spillovers. These changes included the D614G substitution in the spike protein (spikeD614G) and the P323L substitution in the viral RNA-dependent RNA polymerase (NSP12 P323L). SpikeD614G was associated with increased transmissibility and infectivity, while NSP12 P323L was viewed as a potentially coincidental mutation. However, the combination of these two mutations led to the emergence of the B.1 lineage, which has come to dominate the global landscape of SARS-CoV-2.
The role of NSP12 P323L in the virus's biology remained poorly understood, as it was overshadowed by the spike protein mutation. NSP12 is a critical component of the viral replication complex, forming a complex with NSP7 and NSP8. This complex interacts with other viral proteins that alter intracellular membranes' structure. NSP12 itself consists of several domains, including the N-terminal nucleotidyl transferase (NiRAN) domain, the interface domain, and the C-terminal polymerase domain. While the structural analysis identified the NSP12 P323L substitution within the interface domain, its fun
ctional significance was not immediately apparent.
The Study's Hypothesis
The study's primary hypothesis was that the NSP12 P323L substitution is a host adaptation mutation that alters NSP12's interaction with host cell proteins, consequently impacting its function in viral RNA synthesis. Previous research had already highlighted the role of host proteins in interacting with SARS-CoV-2 proteins, suggesting that amino acid substitutions in viral proteins could modulate these interactions and influence the viral life cycle. Given that many RNA-dependent RNA polymerases from various viruses interact with host proteins, including chaperones and co-factors, it was reasonable to assume that NSP12 would follow a similar pattern.
Investigating the Interaction between NSP12 and Host Proteins
To test their hypothesis, the researchers employed a combination of biochemical and virological approaches. They characterized the interaction between NSP1 2P323 and NSP12 L323 and the host cell proteome. The researchers found that both variants of NSP12 interacted with the T-complex protein ring complex (TRiC), a molecular chaperone that plays a crucial role in protein folding and stability. On the other hand, there was a differential association between NSP12 variants and components of a phosphatase complex, including protein phosphatase 2 (PP2A) and STRN3. Notably, the virus expressing NSP12 L323 was less sensitive to perturbations in PP2A, indicating a functional difference between the two variants.
Validation of Interactions and Functional Implications
To confirm the interactions between NSP12 and host proteins, the researchers employed various techniques, including pull-down assays and mass spectrometry. These methods provided quantitative data on the cellular interactome of NSP12 P323 and NSP12 L323. While the interactomes of both variants shared common components, some interactions were enhanced in the presence of NSP12 L323. Notably, STRN3, a regulatory subunit of PP2A, and Epiplakin (EPPK1) exhibited stronger associations with NSP12 L323. Importantly, these interactions were found to be independent of RNA.
Functional experiments were conducted to evaluate the impact of the interactions on SARS-CoV-2 replication. TRiC, a key player in the interactions, was inhibited using a small molecule inhibitor, HSF1A. The results demonstrated that the disruption of TRiC led to a significant reduction in viral RNA, protein abundance, and viral titers. This finding suggested that TRiC played a vital role in maintaining NSP12 stability and function.
The researchers also sought to investigate the biological relevance of the differential interactions with PP2A and STRN3. Depletion of these proteins in infected cells revealed intriguing results. While depletion of STRN3 had minimal impact on SARS-CoV-2 NSP12 P323, depletion of PP2A resulted in a significant reduction in viral RNA, protein abundance, and titers. However, the virus expressing NSP12 L323 proved to be less sensitive to the effects of PP2A depletion.
Discussion
The study's findings provide valuable insights into the intricate interplay between SARS-CoV-2 NSP12 and host cell proteins. While the D614G substitution in the spike protein significantly contributed to the virus's fitness and transmission advantage, the P323L substitution in NSP12 was not merely a bystander mutation. Instead, it appeared to confer a fitness advantage and may represent a host adaptation mutation.
The interactions between NSP12 and host proteins, particularly TRiC, were shown to be crucial for SARS-CoV-2 replication. TRiC's role in protein folding and stability indicated that it played a key part in maintaining NSP12's structure. Disruption of TRiC led to a reduction in viral titers, highlighting the importance of these interactions in the virus's life cycle.
Furthermore, the differential interactions of NSP12 variants with PP2A and STRN3 suggest that the P323L substitution may act as a host adaptation mutation. Similar host-adapting mutations have been observed in other viruses, including influenza. The study's results support the notion that SARS-CoV-2 adaptation in humans is not limited to changes in the spike protein but extends to other viral proteins, emphasizing the complex nature of virus evolution.
In conclusion, the study conducted by the University of Liverpool and the University of Bristol represents a significant contribution to our understanding of SARS-CoV-2 biology and evolution. It highlights the importance of considering the interplay between viral proteins and host cell proteins in the context of virus adaptation, shedding light on the intricate mechanisms that underpin the virus's success in infecting and spreading among human populations. This research underscores the need for continued exploration of SARS-CoV-2's molecular interactions to develop more effective strategies for combatting the ongoing pandemic and future viral threats.
The study findings were published in the peer reviewed Journal of Virology.
https://journals.asm.org/doi/10.1128/jvi.00424-23
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
