COVID-19 Research: Chinese Study Finds That SARS-CoV-2 Nsp15 Protein Suppresses Type 1 Interferon Production!
COVID-19 Research - Nsp15 Inhibits Type 1 Interferon Production. Feb 12, 2023 2 years, 2 months, 3 days, 21 hours, 23 minutes ago
COVID-19 Research: A new research by scientists from Jiangsu University-China has found that the Nsp15 protein of the SARS-CoV-2 virus suppresses Type I Interferon production by inhibiting IRF3 phosphorylation and nuclear translocation.
 Structure Of the SARS-CoV-2 Virus
Structure Of the SARS-CoV-2 Virus
The SARS-CoV-2 virus is a single positive-stranded RNA virus in the family Coronaviridae, genus Betacoronavirus. The viral genome primarily encodes two large open reading frames (ORFs), ORF1a and ORF1b, which are then translated into two large replicase polyprotein precursors (pp1a and pp1b). Papain-like proteinase (Nsp3) and 3C-like proteinase (3CLpro, also known as Nsp5) cleave these two polyproteins into 16 nonstructural proteins. Aside from these nonstructural proteins, sub-genomic RNA of SARS-CoV-2 encodes four structural proteins (spike envelope, membrane, nucleocapsid) and several accessory proteins.
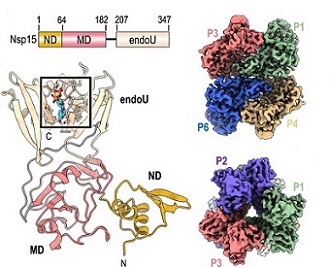
Nsp15 Protein
The Nsp15 protein is a conserved uridine-specific endoribonuclease, and is an integral component of the coronavirus replicase-transcriptase complex (RTC) that processes viral RNA to evade detection by host innate immune systems.
https://pubmed.ncbi.nlm.nih.gov/28158275/
https://pubmed.ncbi.nlm.nih.gov/34204549/
5′-PolyU-containing, negative-sense (PUN) RNAs as viral pathogen-associated molecular patterns (PAMPs) could be recognized by RLRs (RIG-I-like receptors) to activate the IFN response.
However, the poly-U specific endonuclease (EndoU) of coronavirus activity cleaves PolyU sequences from PUN RNAs to evade the innate immune system.
https://pubmed.ncbi.nlm.nih.gov/32198201/
In addition, past
COVID-19 Research has shown that SARS-CoV-2 Nsp15 was also found to inhibit de novo autophagy induction.
https://pubmed.ncbi.nlm.nih.gov/33974846/
Past studies have shown that SARS-CoV-2 Nsp15 could antagonize the host innate immune response, but the underlying mechanism remains to be determined.
Inhibition Of Interferon (IFN) Production
The SARS-CoV-2 virus like other coronaviruses, has evolved many strategies to inhibit interferon (IFN) production.
The study team identified that SARS-CoV-2 Nsp15 suppresses IFN production by antagonizing the RLR-mediated antiviral signaling pathway. Overexpressed Nsp15 obviously reduced mRNA levels of IFN-β and IFN-stimulated genes (ISG56, CXCL10), and also inhibited IRF3 phosphorylation and nuclear translocation.
Mechanically, the poly-U-specific endonuclease domain (EndoU) of Nsp15 directly associates with the kinase domain (KD) of TBK1, thereby Nsp15 interfered TBK1 interacting with IRF3 and reduc
ed TBK1-mediated IRF3 phosphorylation. In addition, Nsp15 also prevented nuclear translocation of phosphorylated IRF3 via binding and reducing the nuclear import adaptor karyopherin α1 (KPNA1) protein.
In this study, the researchers report that Nsp15 suppresses IFN production in two different ways:
-Nsp15 weakened the interaction between TBK1 and IRF3 by competitively combining with TBK1, thereby reducing IRF3 phosphorylation;
-Nsp15 inhibited nuclear translocation of phosphorylated IRF3 via binding and reducing karyopherin α1 (KPNA1) protein. To our knowledge, this is the first time to clarify the mechanism of Nsp15 antagonizing the innate immune pathway in detail. This study will provide an optional target for drug development against COVID-19 disease
This study findings reveal the novel mechanism of Nsp15 antagonizing the innate immune pathway and provided an optional target for drug development against COVID-19 disease.
The study findings were published on a preprint server and are currently being peer reviewed.
https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4345044
Typically, during viral infection, type I IFN responses act as the first line of antiviral immunity to inhibit viral proliferation. Clinical symptoms in COVID-19 patients and studies in animal models or human cells have shown that SARS-CoV-2 weakly induces type I IFN response, suggesting that it may have evolved a more efficient way of surviving in the host.
This new study confirmed the inhibitory effect of Nsp15 on RIG-I-induced type IFN and clarified two potential regulatory mechanisms of SARS-CoV-2 Nsp15 in blocking the RLR-signaling pathway.
TBK1 plays an essential role in cell signaling pathway activation, pathogen autophagy induction, and cell cycle maintenance.
As a key component of the RIG-I signaling pathway, TBK1 is also critical for inducing IFN production.
Many viruses have evolved numerous mechanisms to inhibit IFN production by targeting TBK1.
For example, Zika virus NS5 protein reduced IFN expression by inhibiting TBK1 activation and IRF3 phosphorylation.
https://pubmed.ncbi.nlm.nih.gov/30530224/
This new study finds that SARS-CoV-2 Nsp15 interacts with TBK1, and the KD domain of TBK1 was indispensable for their interaction. The KD domain is the key enzyme activity region of TBK1. Especially autophosphorylation at Ser-172 in the KD domain can activateTBK1 kinase, and is an essential step for virus-triggered signaling.
In this research, the interaction between Nsp15 and the KD domain of TBK1 neither masked the Ser-172 phosphorylation, nor blocked TBK1 activity, but significantly reduced IRF3 phosphorylation.
Subsequent research testified that Nsp15 weakens the interaction between TBK1 and IRF3 by competitively combining with TBK1, thereby reducing IRF3 phosphorylation, which has also been observed in SARS-CoV-2 ORF6-mediated interferon regulation.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7501843/
The Nsp15protein is a nidoviral RNA uridylate-specific endoribonuclease (NendoU) carrying the C-terminal catalytic domain that is conserved in coronaviruses, toroviruses, and arteriviruses.
Past studies have shown that coronavirus Nsp15 is required for evasion of dsRNA sensors.
https://pubmed.ncbi.nlm.nih.gov/28484023/
It has been shown that the loss of Nsp15 enzyme activity in porcine epidemicdiarrhea virus (PEDV) activated interferon responses and reduced viral titers.
https://pubmed.ncbi.nlm.nih.gov/30728254/
The study findings demonstrated that SARS-CoV-2 Nsp15 interacts with TBK1 via its EndoU domain (E) but not ND and MD domains.
Interestingly, ME and E truncations alone inhibited TBK1-induced IFN expression by 44.8% and 67.2%, respectively. It indicated that the EndoU domain of Nsp15 played an important role in regulating the RIG-I signaling pathway.
It was also found that overexpression of Nsp15 also inhibited IRF3-induced IFN production and IRF3 nuclear translocation.
The study team speculates that Nsp15 may also target downstream proteins of IRF3. Karyopherins (KPNAs) participate in the transport of many cytokines by recognizing specific nuclear localization signals or nuclear export signals, many of which are transcription factors involved in cell growth and differentiation, as well as cytokines regulating host immunity.
https://pubmed.ncbi.nlm.nih.gov/15450977/
Numerous viruses suppress innate antiviral responses to promote viral replication by targeting KPNAs.
The SARS-CoV-2 ORF6 also interacted directly with the Nup98-Rae1 complex, impairing karyopherin complex docking and blocking STAT1 and STAT2 nuclear import, and subsequently impairing transcriptional induction of IFN-stimulated genes (ISGs).
https://pubmed.ncbi.nlm.nih.gov/33097660/
The study findings showed a potential mechanism that SARS-CoV-2 Nsp15 hijacked KPNA1 to block IRF3 nuclear imports, leading to suppression of both IFN production and following IFN-stimulated gene expression. Moreover, Nsp15 decreased KPNA1 expression in a dose-dependent manner. These study findings provided new insights into SARS-CoV-2 Nsp15 preventing IRF3 nuclear translocation.
The study team concluded that the study identified SARS-CoV-2 Nsp15 as an important antagonist of type I IFN via two interfering mechanisms on the RLR-signaling pathway.
The study findings provide new insights into SARS-CoV-2 escaping the host antiviral innate immunity and optional drug target for the treatment of COVID-19 disease.
For the latest
COVID-19 Research, keep on logging to Thailand Medical News.
