COVID-19 Research: University Of Texas MD Anderson Cancer Center Detailed Interactome Study Between SARS-CoV-2 and Host Cells Unveils 437 Proteins
Source: COVID-19 Research Jan 05, 2021 4 years, 3 months, 3 weeks, 10 hours, 3 minutes ago
COVID-19 Research: Scientists from the University of Texas MD Anderson Cancer Center in Houston in a new detailed interactome study between the SARS-CoV-2 coronavirus and human host cells have identified 437 human proteins as high-confidence interacting proteins.
.jpg)
Host-virus protein-protein interaction is the key component of the SARS-CoV-2 lifecycle.
The study team conducted a comprehensive interactome study between the virus and host cells using tandem affinity purification and proximity labeling strategies and identified 437 human proteins as the high-confidence interacting proteins. Functional characterization and further validation of these interactions elucidated how distinct SARS-CoV-2 viral proteins participate in its lifecycle, and discovered potential drug targets to the treatment of COVID-19.
The interactomes of two key SARS-CoV-2 encoded viral proteins, NSP1 and N protein, were compared with the interactomes of their counterparts in other human coronaviruses. These comparisons not only revealed common host pathways these viruses manipulate for their survival, but also showed divergent protein-protein interactions that may explain differences in disease pathology. This comprehensive interactome of coronavirus disease-2019 provides valuable resources for understanding and treating this disease.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2020.12.31.424961v1
Simply by taking into consideration the transmission, morbidity and mortality patterns of the COVID-19 pandemic, it is evident that SARS-CoV-2 is a pathogenic coronavirus that spreads easily through the air.
While the mass vaccination programs kicks off in many countries of the world, the search for viable antiviral treatments, drugs and therapeutics.
Significantly one of the key facets of SARS-CoV-2 lifecycle is host-virus protein-protein interactions. As these may unveil promising drug targets, the analysis of host-virus interactome (i.e., a complete set of molecular interactions in a particular cell) is critically needed and have actually been reported recently.
The study team had two well-established strategies for studying protein-protein interactome on their disposal ie affinity purification and a proximity labeling-based strategy, which is followed by mass spectrometry analysis.
It was recently shown that the combination of these methods is the optimal way to generate comprehensive insights.
The study team led by Dr Zhen Chen from the Department of Experimental Radiation Oncology at the University of Texas MD Anderson Cancer Center in Houston (USA) started with a quest for key human proteins that are implicated in the SARS-CoV-2 life cycle.
The team applied the two aforementioned strategies: tandem affinity purification with the SFB (S-protein, FLAG epitope, and streptavidin-binding peptide) tag, as well as proximity labeling by using a second-generation biotin ligase, BioID2.
Detailed genome annotation disclosed 29 SARS-CoV-2 gene products including 16 non-structural proteins, 4 structural protein
s and 9 accessory factors. Furthermore, stable viral gene expression was done in cells and subsequently verified by immunoblotting technique.
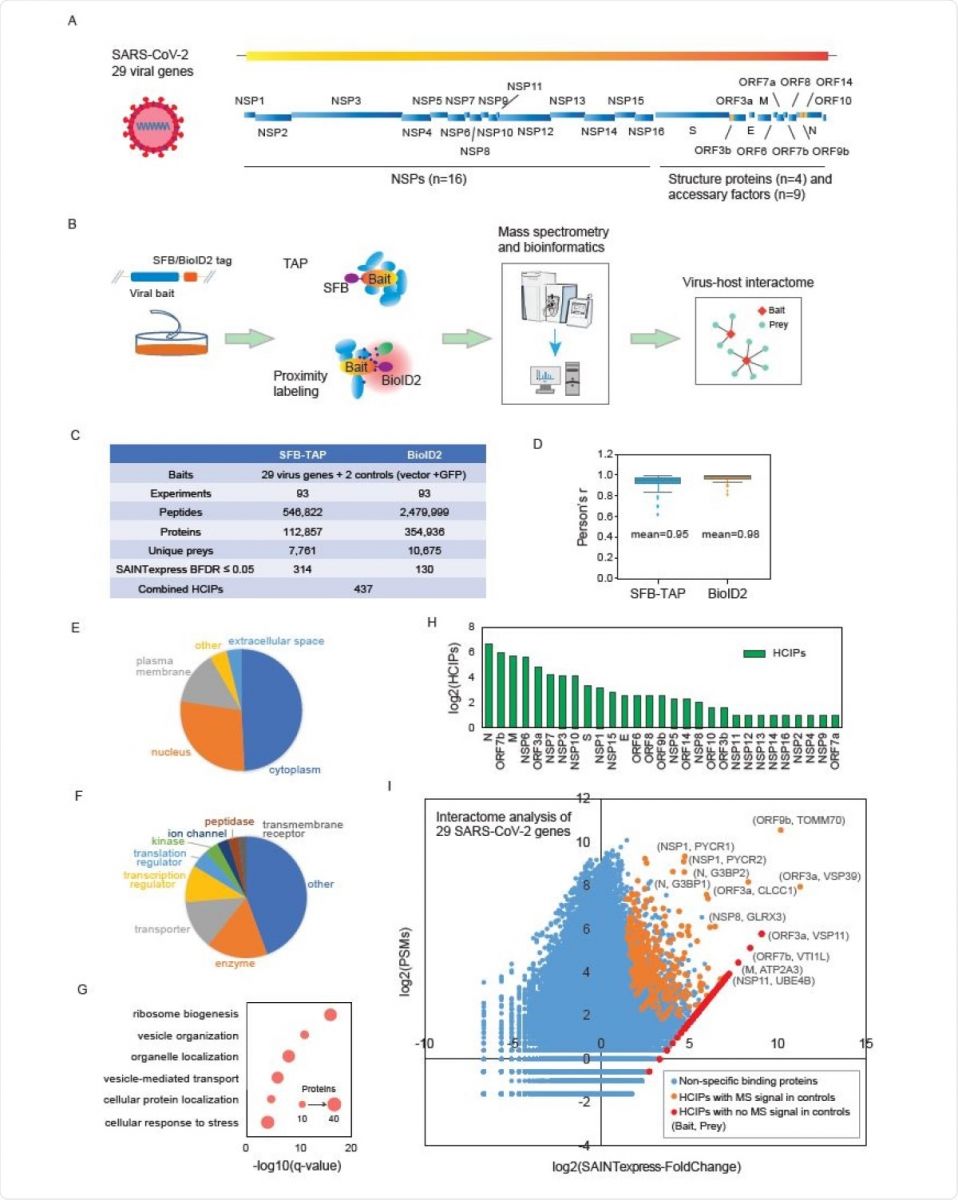 Details of the SFB-TAP and BioID2 interactome experiments. (A) SARS-CoV-2 genome annotation, predicting 29 virus gene products. The 16 non-structure proteins (NSPs) are cleaved products of the large polyprotein open reading frame (ORF)1ab or ORF1a. These polyproteins are cleaved into small function fragments or NSPs after translation. (B) Workflow for the comprehensive virus-host interactome analysis. Two different labeling strategies, SFBTAP and BioID2 labeling, were applied in the study. Samples were analyzed by Q Exactive HF mass spectrometry (MS). (C) Summary of the datasets obtained from SFB-TAP and BioID2 results, including the number of high-confidence interacting proteins (HCIPs). BDFR, Bayesian false discovery rate. (D) Pearson correlation coefficient among three independent biological replicates of the SFB-TAP results and the BioID2 labeling experiments. (E-G) GO analysis. GO enrichment was performed using Ingenuity Pathway Analysis. Protein localization (E), molecular function (F), and biological function (G) are plotted in a single panel. (H) HCIPs identified in the purification of each SARS-CoV-2 gene. (I) Correlation between peptide-spectrum matches (PSMs) of identified proteins and their fold change calculated by SAINTexpress.
Details of the SFB-TAP and BioID2 interactome experiments. (A) SARS-CoV-2 genome annotation, predicting 29 virus gene products. The 16 non-structure proteins (NSPs) are cleaved products of the large polyprotein open reading frame (ORF)1ab or ORF1a. These polyproteins are cleaved into small function fragments or NSPs after translation. (B) Workflow for the comprehensive virus-host interactome analysis. Two different labeling strategies, SFBTAP and BioID2 labeling, were applied in the study. Samples were analyzed by Q Exactive HF mass spectrometry (MS). (C) Summary of the datasets obtained from SFB-TAP and BioID2 results, including the number of high-confidence interacting proteins (HCIPs). BDFR, Bayesian false discovery rate. (D) Pearson correlation coefficient among three independent biological replicates of the SFB-TAP results and the BioID2 labeling experiments. (E-G) GO analysis. GO enrichment was performed using Ingenuity Pathway Analysis. Protein localization (E), molecular function (F), and biological function (G) are plotted in a single panel. (H) HCIPs identified in the purification of each SARS-CoV-2 gene. (I) Correlation between peptide-spectrum matches (PSMs) of identified proteins and their fold change calculated by SAINTexpress.
Importantly the identified proteins were filtered with the use of Significance Analysis of INTeractome (abbreviated as SAINTexpress). The study team had also built an interaction network by utilizing the 437 identified virus-host protein-protein interactions, which enabled all the complex analyses that they have pursued.
Besides a total of 437 high-confidence interacting proteins that bind to one or more SARS-CoV-2 genes, the researchers have also identified several gene products, M protein, NSP6, ORF3a, ORF6 and ORF7b that interacted with host cell membrane proteins and complexes.
Significantly the transmembrane domain prediction also indicated that these viral gene products contain at least one transmembrane domain in their protein sequences with the exception of ORF6, which is actually a short protein with only 61 amino acids.
Also 314 high-confidence interacting proteins were identified from tandem affinity purification experiments, while 130 of them with the use of BioID2 strategy. Interestingly, only seven proteins overlapped between these two methods.
Interestingly when the interactomes of NSP1 and N protein (i.e., two key SARS-CoV-2 proteins) were compared with other human coronaviruses, host pathways manipulations and divergent protein-protein interactions responsible for differences in disease pathology were uncovered.
M protein, NSP6, ORF3a, ORF6, and ORF7b help viral infection, trafficking, and maybe the budding of the virion; NSP1, NSP3, NSP5, NSP6, NSP7, and N protein suppress host cell replication, transcription, and translation, and at the same time contribute to the same processes in the viral lifecycle; S protein facilitates the formation of new viruses after viral replication and translation of structural proteins; NSP1, NSP5, N protein, M protein, and ORF7 may inhibit host cell antiviral responses and therefore promote the survival of the infected virus, and they also prolong host cell survival to increase viral production. NSP6 and NSP7 may manipulate host cell metabolism and signaling transduction pathways. Understanding in detail how the virus and host cell communicate will help researchers to find or design therapeutic strategies to suppress viral infection
The systemic study of the SARS-CoV-2 protein-protein interaction network provides useful data on viral gene/protein functions and potential underlying mechanisms, which could lead to the identification of new drug targets for the treatment of COVID-19, according to the study team.
Importantly the obtained interactome dataset not only proved some previously established host-virus interactions, but also uncovered manifold novel interacting proteins that may be pivotal for all relevant components of SARS-Cov-2 lifecycle.
The study findings will undoubtedly be exploited in the ongoing fight against COVID-19; however, these datasets may also suggest new ways to combat any potential new emerging coronavirus diseases in the future, which is continuously a threat.
For the latest
COVID-19 Research, keep on logging to Thailand Medical News.
.jpg)
