Deciphering The Intricacies Of COVID-19 Progression - Unraveling The Role Of Natural Killer Cell-Mediated Cytotoxicity
Nikhil Prasad Fact checked by:Thailand Medical News Team Feb 29, 2024 1 year, 10 months, 1 week, 2 days, 13 hours, 3 minutes ago
COVID-19 News: The COVID-19 pandemic has presented a daunting challenge to global public health, characterized by a broad spectrum of clinical manifestations ranging from asymptomatic infection to severe respiratory distress and death. Despite intense research efforts, the underlying molecular and cellular mechanisms driving the variability in disease severity remain incompletely understood. However, recent studies have shed light on the pivotal role of host immune responses in shaping the clinical trajectory of COVID-19.
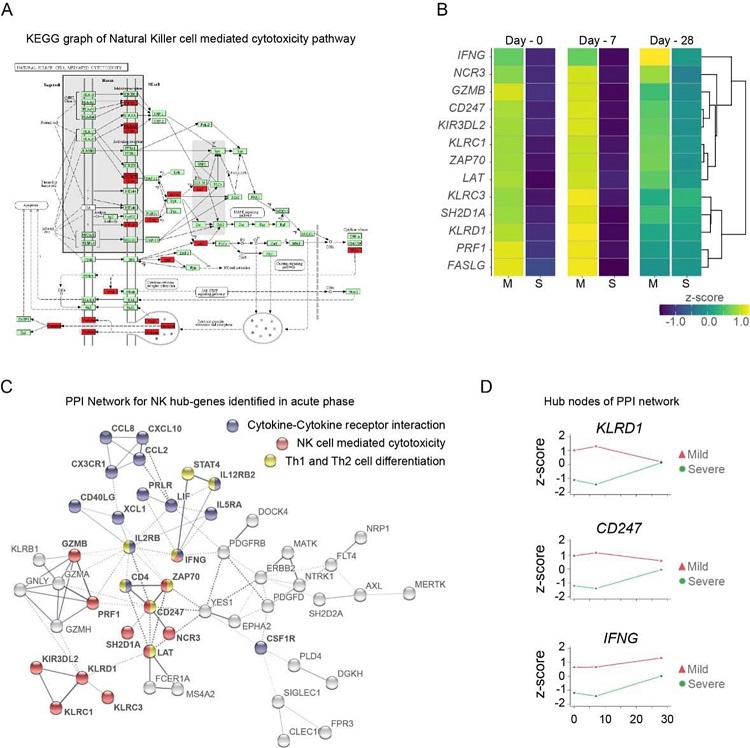 Gene function network for Natural Killer cell hub-genes with differential expression levels between mild and severe patients during acute phase of COVID-19. (A) KEGG pathway of Natural Killer cell mediated cytotoxicity represents the set NK cell hub-genes upregulated (red-boxes) in mild versus severe patients. The green boxes correspond to genes without differential expression. (B) Heatmap shows the differential expression levels of NK cell hub-genes over time (Day-0, 7, and 28 after recruitment) separated by mild and severe groups. The expression levels are represented by the z-score of normalized counts. Dendrogram shows the hierarchical clustering of genes. (C) Protein-protein interaction (PPI) network for upregulated genes during the acute phase in mild patients. The network corresponds to the principal clusters with more interaction between proteins and highlight the three most represented pathways: Cytokine-cytokine receptor interaction (blue); NK cell mediated cytotoxicity (red); and Th1 and Th2 cell differentiation (yellow). (D) Time course expression levels for the main protein nodes identified in PPI network during the acute phase of COVID-19. The trajectories of these genes are graphed as days after recruitment (0, 7, and 28 days) for mild (red triangle) and severe (green circle) groups and their enrichment is represented by the z-score of normalized counts.
Gene function network for Natural Killer cell hub-genes with differential expression levels between mild and severe patients during acute phase of COVID-19. (A) KEGG pathway of Natural Killer cell mediated cytotoxicity represents the set NK cell hub-genes upregulated (red-boxes) in mild versus severe patients. The green boxes correspond to genes without differential expression. (B) Heatmap shows the differential expression levels of NK cell hub-genes over time (Day-0, 7, and 28 after recruitment) separated by mild and severe groups. The expression levels are represented by the z-score of normalized counts. Dendrogram shows the hierarchical clustering of genes. (C) Protein-protein interaction (PPI) network for upregulated genes during the acute phase in mild patients. The network corresponds to the principal clusters with more interaction between proteins and highlight the three most represented pathways: Cytokine-cytokine receptor interaction (blue); NK cell mediated cytotoxicity (red); and Th1 and Th2 cell differentiation (yellow). (D) Time course expression levels for the main protein nodes identified in PPI network during the acute phase of COVID-19. The trajectories of these genes are graphed as days after recruitment (0, 7, and 28 days) for mild (red triangle) and severe (green circle) groups and their enrichment is represented by the z-score of normalized counts.
In a new comprehensive investigation covered in this
COVID-19 News report, researchers from leading institutions including Universidad San Sebastián-Chile, Universidad de Concepción-Chile, Hospital Dr. Eduardo Schütz Schroeder-Chile, Hospital Base San José-Chile, Universidad Austral-Chile, and Icahn School of Medicine at Mount Sinai, New York-USA, embark on a journey to decipher the intricate interplay between the immune system and SARS-CoV-2, the virus responsible for COVID-19. Through a meticulous analysis of longitudinal transcriptional changes in peripheral immune cells, the study aims to uncover the critical molecular pathways underlying COVID-19 progression, with a particular focus on the role of genes associated with the natural killer (NK) cell-mediated cytotoxicity pathway.
Unraveling Clinical Heterogeneity: From Asymptomatic Carriers to Severe Cases
The clinical spectrum of COVID-19 is strikingly diverse, with some individuals exhibiting no symptoms while others develop severe respiratory complications nec
essitating intensive care support. Understanding the factors driving this heterogeneity is paramount for developing effective diagnostic tools and targeted therapies. Previous research has identified biomarkers associated with COVID-19 severity, including an imbalance in immune cell populations, lymphopenia, myeloid dysfunction, and T cell activation/exhaustion.
The gene expression profiles differed notably between mild and severe COVID-19 patients. For instance, ICU patients showed higher expression levels of MMP9, S100A8/A9, PADI2, and IL18Rap on day-0 compared to mild or control patients. Conversely, IFNG, CCL2, and CXCL10 cytokines, previously associated with severe illness, exhibited lower expression in ICU patients compared to mildly ill patients as the disease progressed.
These findings underscore the critical role of the immune system in shaping disease outcomes.
Longitudinal Investigation: Design and Methodology
To elucidate the dynamic immune responses during the early stages of COVID-19, the researchers implemented a longitudinal study design. Peripheral blood samples were collected from COVID-19 patients at multiple time points post-diagnosis, allowing for a comprehensive analysis of transcriptome profiles in peripheral blood mononuclear cells (PBMCs). The study meticulously selected samples from both mild and severe cases, enabling a comparative analysis of gene expression patterns over time.
Unveiling Transcriptional Changes: The Role of NK Cell-Mediated Cytotoxicity
The analysis of longitudinal transcriptional changes revealed significant variations in gene expression profiles between mild and severe COVID-19 patients, particularly during the acute phase of infection. Remarkably, genes associated with NK cell-mediated cytotoxicity emerged as central players in distinguishing between mild and severe disease progression. Key genes including KLRD1, CD247, and IFNG exhibited differential expression patterns, highlighting the critical role of NK cells in mounting an effective immune response against SARS-CoV-2.
The role of Natural Killer (NK) cells in COVID-19 progression is significant, with various genes involved in NK cell-mediated cytotoxicity pathways playing key roles. These pathways include genes such as KLRC1, KLRC3, KLRD1, KIR3DL2, and NCR3 receptors, along with effectors like SH2D1A, PRF1, GZMB, FASLG, ZAP70, IFNG, CD247, and LAT. These genes are crucial for regulating cytotoxicity and attracting NK cells to control viral infections. We observed a dynamic transcriptomic profile of NK cell genes, showing higher expression levels in individuals with mild disease compared to severe cases during the early stages of infection. However, expression levels of these NK cell genes became more similar between mild and severe patients as the disease progressed. Additionally, we noted an up-regulated gene signature consistent with dominant neutrophil activities in severe cases, even after recovery. The NK cell gene hub includes activating (KLRC3, NCR3) and inhibitory (KLRC1, KIR3DL2) genes, as well as regulatory and effector proteins (KLRD1, GZMB, PRF1), which contribute to a well-coordinated NK cell activity profile. The KRLD1 gene, encoding the CD94 protein, plays a central role in regulating NK cell cytotoxicity by balancing activating and inhibitory functions.
Connecting the Dots: Gene Co-expression Networks and Immune Pathways
To unravel the intricate connections within the immune system, the researchers employed a weighted gene correlation network analysis. This approach identified three co-expressed gene modules - blue, brown, and turquoise - that correlated with mild and severe COVID-19 patients. These modules revealed distinct pathways associated with T-cell activation, platelet function, neutrophil activation, and inflammatory responses. The findings underscored the relevance of Th1/Th2 cell differentiation pathways, emphasizing their role in mild COVID-19 progression.
Integrating Innate and Adaptive Immunity: Implications for Disease Severity
A holistic interpretation of the findings highlighted the critical interplay between innate and adaptive immune responses in shaping COVID-19 outcomes. The orchestrated transcriptional response of dominant NK cell activities in mild patients underscored the importance of early innate immune responses in mitigating disease severity. Furthermore, the integration of transcriptomic analyses with clinical outcomes offered valuable insights into potential diagnostic markers and therapeutic targets.
Conclusion: Towards Precision Medicine in COVID-19 Management
In conclusion, this groundbreaking study provides a comprehensive understanding of the molecular landscape underlying COVID-19 progression. By unraveling the intricate interplay between host immune responses and viral pathogenesis, the study offers valuable insights into potential diagnostic markers and therapeutic targets. The identification of key molecular pathways, particularly those associated with NK cell-mediated cytotoxicity, paves the way for the development of precision medicine strategies to identify high-risk patients and engineer effective treatments against SARS-CoV-2 and related respiratory viruses.
This research not only advances our understanding of COVID-19 pathogenesis but also holds promise for personalized medicine approaches in infectious diseases. By elucidating the molecular mechanisms driving disease severity, researchers are poised to develop targeted interventions that could revolutionize the management of COVID-19 and improve clinical outcomes worldwide.
The study findings were published in the peer reviewed journal: eLife.
https://elifesciences.org/reviewed-preprints/94242
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
