French Researchers Uncover That SARS-CoV-2 Nsp3 Protein Interacts With Human Host RNA G-Quadruplexes, Contributing To Virulence
Source: SARS-CoV-2 Research Jul 27, 2021 4 years, 5 months, 2 days, 15 hours, 16 minutes ago
A new French study led by the Institut Pasteur has found that a domain of the SARS-CoV-2 coronavirus Nsp 3protein interacts with unusual human host deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) structures called G-quadruplexes or G4s and this interaction could possibly be contributing to virulence. The use of inhibitors of these interactions could serve as potential antiviral compounds.
The SARS-CoV-2 genome contains 14 open reading frames, with ORF1a and ORF1b occupying two thirds of the genome and encoding 16 non-structural proteins (Nsp1-16) after auto-proteolytic processing of the polyproteins.
The
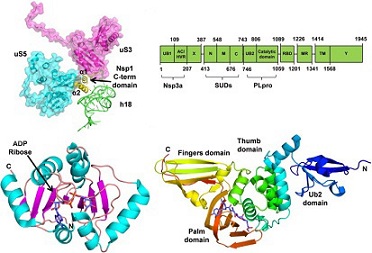
structural protein Nsp3 plays a crucial role in the formation and activity of the viral replication/transcription complex. Originally, this protein was shown to contain a non-conserved domain called SARS-Unique Domain or SUD, which is absent in less pathogenic coronaviruses infecting humans or incomplete in the MERS-CoV Nps3 protein.
The detailed 3D-structure of this domain has revealed the presence of two macrodomains (SUD-N and SUD-M) and a frataxin-like domain (SUD-C), corresponding to the N-terminal, Middle and C-terminal parts of SUD, respectively. These three domains are called SUD-N, SUD-M and SUD-C.
G4s are non-canonical nucleic acid structures formed by G-rich RNA or DNA sequences, which result from the stacking of two or more guanine quartets stabilized by a central spine of cations. These structures are highly polymorphic and have been extensively studied
in vitro, using biophysical and structural approaches. G4s regulate major eukaryotic cell processes and are enriched in key regulatory regions of their genomes and transcriptomes including telomeres, promoters and 5′UTR of highly transcribed genes. While DNA G4s regulate the transcription of a number of human genes, especially oncogenes, RNA G4s participate in several mechanisms linked with mRNA metabolism, such as their translation, splicing, stability, polyadenylation and localization. G4s are also present in the genomes of DNA and RNA viruses and control critical steps of their replication. For instance, G4s present in the HIV-1 genome regulate reverse transcription and transcription steps and these regulations require their interaction with cellular proteins. Conversely, some viral proteins act on viral replication through their interaction with RNA G4s.
SARS-CoV SUD N and M macrodomains have homologous structures and both interact with DNA and RNA oligonucleotides, especially those folded into guanine-quadruplex (G4) structures.
SUD-NM/G4 structural models of this complex have been used to identify the amino-acids of SUD putatively involved in G4 binding and to study the conformational dynamics of the SUD-NM/G4 complex. At a functional level, using a reverse-genetic system, it has been shown that SUD-M deletion and targeted mutations altering G4 binding largely affect the activity of the SARS-CoV replication/transcription complex.
SARS-CoV SUD also interacts with cellular proteins. A fusion of SUD and PLpro Nsp3 domains binds to the E3 ubiquitin ligase RCHY1 and this interaction contributes to the virulence of SARS-CoV in human cells. More recently, it has been shown that both SARS-CoV and SARS-CoV-2 SUD domains interact with the human poly(A)-binding protein (PABP)-interacting protein A (Paip1) and that this interac
tion enhances SARS-CoV RNA translation.
The SARS-CoV SUD domain also modulates the inflammatory response in lung epithelial cells.
Altogether, these data strongly support the role of SARS-CoV SUD in viral replication and cell response to infection and suggest that these effects may be mediated in part by interactions of SUD with G4 structures.
The study findings were published in the peer reviewed journal:
Nucleic Acids.
https://academic.oup.com/nar/article/49/13/7695/6316836#272508508
The study also demonstrated that G4 ligands, the chemical that binds with G4s, prevent this SUD-NM/G4 interaction. The study results could pave the way for developing inhibitors of these interactions as potential antiviral compounds.
SARS-CoV-2
The novel coronavirus: SARS-CoV-2, the virus behind COVID-19 pandemic, continues to spread across the globe, infecting almost over 195 million people. The pandemic has claimed more than 4.2 million lives.
The SARS-CoV-2 virus replication relies on a series of interactions between viral proteins and various cellular partners like nucleic acids. Therefore, investigating these interactions is essential to showcase the process of viral replication.
The researchers from Institut Pasteur, the Ecole Polytechnique, the Institut Curie, Inserm, the CNRS, and the universities of Paris, Paris-Saclay, Bordeaux, and Toulouse decided to investigate whether a SUD domain is equally present in SARS-CoV-2 Nsp3. If this domain was present, the team aimed to analyze its ability to bind G4s and to see whether these interactions can be modulated by G4-ligands.
The findings showed that the SARS-CoV-2 Nsp3 protein contains a SUD domain that interacts with specific DNA and RNA G4s but not with the putative G4s predicted in the SARS-CoV-2 genome.
The study team also found that these interactions can be disrupted by mutations preventing oligonucleotides from folding into G4 structures and by G4 ligands.
Importantly host-virus interactions represent promising targets for antiviral strategies. The study focused on the interaction between SARS-CoV-2’s SUD domain Nsp3 protein and the DNA or RNA G-quadruplexes. The interactions reflect a high therapeutic potential.
The research findings pave the way for further studies on the role of the SUD/G4 interactions during SARS-CoV-2 replication. It shows promise in the development of preventive therapeutics that can hinder SARS-CoV-2 reproduction.
Various research are ongoing to determine the preferential partners of the SARS-CoV-2 SUD domain, more likely messenger RNAs (mRNAs) coding for proteins involved in the immune response, stress response, or inflammation.
Corresponding author, Dr Marc Lavigne from the Institut Pasteur, Département de Virologie told Thailand Medical News, “Our results, showing the capacity of some G4-ligands to inhibit the SARS-CoV-2 SUD/G4 interaction paves the way for a global screening of molecules able to inhibit this interaction and to test their antiviral properties.”
The study team also developed G4 ligands and demonstrated that these ligands, which hinder interactions between the SARS-CoV-2 protein and the G4 structure, elicit an antiviral activity. Hence, these structures could be used in therapeutics to combat the current pandemic.
Although over 3.91 billion vaccine doses have been administered globally, many countries still report surging cases brought about by emerging SARS-CoV-2 variants.
Hence, apart from vaccines, it is extremely critical to develop therapeutics to prevent and treat COVID-19.
For the latest
COVID-19 Research, keep on logging to Thailand Medical News.
