French Study Claims That SARS-CoV-2 Adapts to Human Cells by Hijacking RNA Processes
Nikhil Prasad Fact checked by:Thailand Medical News Team Oct 30, 2024 1 year, 3 months, 3 days, 19 hours, 26 minutes ago
Medical News: COVID-19 Virus Exploits Human RNA Machinery for Survival, New Study Reveals
Researchers at the University of Montpellier and Institut de Recherche en Infectiologie de Montpellier (IRIM) in France uncover how SARS-CoV-2 exploits human cells’ RNA structures to increase its replication efficiency.
 French Study Claims That SARS-CoV-2 Adapts to Human Cells by Hijacking RNA Processes
French Study Claims That SARS-CoV-2 Adapts to Human Cells by Hijacking RNA Processes
Scientists have recently revealed that SARS-CoV-2, the virus causing COVID-19, has adapted in ways that allow it to exploit the human cell's genetic machinery. Unlike other viruses, SARS-CoV-2 relies on a unique strategy that involves altering RNA processes within infected cells, making them more efficient at producing viral components. The discovery sheds light on why the virus has been so adept at spreading in humans and opens the door for potential new treatments.
Uncovering the Virus’s Strategy to Hijack RNA
Viruses like SARS-CoV-2 cannot replicate on their own and rely on host cells to create new virus particles. The study, published by scientists from the University of Montpellier and IRIM, examines how SARS-CoV-2 uses RNA modifications to hijack the human cell’s systems. This
Medical News report explains the findings, highlighting that while human cells use certain genetic sequences called codons efficiently, SARS-CoV-2 appears to have developed a different tactic to thrive.
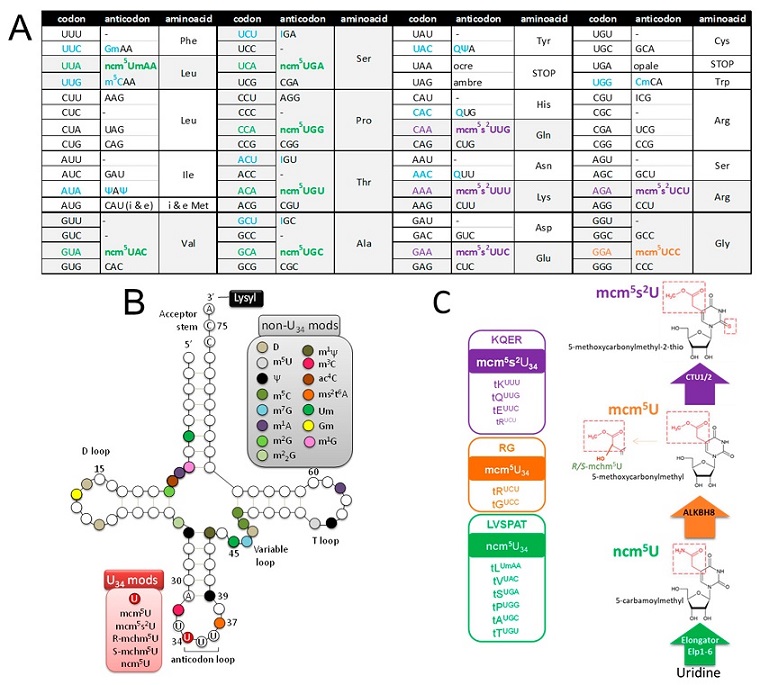
Using a technique called codon bias analysis, the researchers discovered that SARS-CoV-2 uses “suboptimal” codons, meaning the genetic sequences it relies on do not match those that human cells naturally prefer. Instead, SARS-CoV-2 forces the human cell to modify its RNA structure in a way that allows these “suboptimal” codons to be read efficiently. The virus achieves this by changing the host’s transfer RNA (tRNA), which is crucial for assembling proteins. This manipulation of tRNA, specifically at a site known as the U34 wobble position, is essential for efficient viral replication and opens new doors to understand SARS-CoV-2's adaptability and transmission.
The Importance of RNA in Viral Replication
In normal human cells, genetic information is copied into RNA, which then helps produce proteins required for cell functions. Each three-base segment in RNA, known as a codon, corresponds to a specific amino acid or building block of proteins. The researchers found that SARS-CoV-2 has a distinct preference for certain codons - those rarely used by human cells. This mismatch should, in theory, slow down the virus’s ability to replicate. However, SARS-CoV-2 compensates for this inefficiency by altering the structure of specific tRNA molecules at the U34 position, which the virus relies on heavily.
Without these changes, SARS-CoV-2 would struggle to replicate efficiently, suggesting that these tRNA modifications are vital to the virus's lifecycle. According to the researchers, this codon bias and the resulting modifications in the human cell’s
translation process highlight the virus’s unique mechanism for replication and survival.
What Codon Bias Means for SARS-CoV-2’s Evolution and Transmission
Codon bias refers to the tendency of an organism to use certain codons over others. In human cells, commonly used codons are paired with a ready supply of corresponding tRNA, allowing for quick and efficient protein production. Viruses that adapt to match their host’s codon preferences are typically more efficient at using the host’s cellular machinery.
However, SARS-CoV-2 diverges from this pattern. The study found that the virus prefers “A-ending” codons, which are underrepresented in the human genome. This preference could theoretically hinder the virus’s ability to replicate within human cells. But by manipulating the tRNA epitranscriptome - the pool of tRNA molecules in human cells - SARS-CoV-2 manages to override this disadvantage. Specifically, the virus induces modifications to the U34 wobble position on certain tRNAs, making it easier to read its preferred codons. This adaptation ensures that viral proteins are efficiently produced, thus supporting rapid viral replication and enhancing infectivity.
How SARS-CoV-2 Adjusts Human Cells to Meet its Needs
The study found that certain viral codons, like LysAAA, GlnCAA, GluGAA, and ArgAGA, require specific modifications on the human tRNA at the U34 wobble position for efficient translation. To support the virus's needs, human cells infected with SARS-CoV-2 showed increased levels of U34 tRNA modifications. Without these modifications, SARS-CoV-2 would face obstacles in replicating effectively, as the host cell would struggle to interpret its unique codon sequences correctly. This discovery provides evidence that the virus has evolved a unique method of manipulating the host cell's RNA machinery to favor its survival.
To further confirm this finding, the research team tested human cells with altered tRNA modifications. Cells with reduced U34 modifications struggled to support SARS-CoV-2 replication, indicating that these RNA changes are essential for the virus. The researchers used advanced mass spectrometry to detect and measure these tRNA modifications, supporting the hypothesis that SARS-CoV-2 depends on a modified host RNA environment for efficient replication.
Implications for Future Research and Potential Treatments
Understanding the specific molecular mechanisms that SARS-CoV-2 uses to manipulate tRNA modifications could open new avenues for COVID-19 treatments. By targeting the virus’s ability to induce these RNA changes, researchers may develop therapies that inhibit SARS-CoV-2 replication without harming the host cells.
The study also raises questions about the potential for other viruses to use similar strategies, suggesting that further research on virus-host interactions involving RNA modifications is warranted. According to the authors, this insight into the virus's adaptation mechanism could pave the way for antiviral strategies aimed at disrupting tRNA modification processes, thereby hindering the virus's ability to hijack the human cell machinery.
Study Conclusions
The study concludes that SARS-CoV-2’s ability to manipulate the host cell's RNA modification processes is a significant factor in its rapid transmission and adaptability. The research highlights the need for further studies to explore the full impact of viral-induced RNA modifications on human health. The virus's reliance on human tRNA modifications, particularly at the U34 wobble position, could present a novel target for antiviral therapies. Targeting these RNA processes could disrupt the virus’s replication cycle, offering a new strategy in the fight against COVID-19 and potentially other RNA viruses that might use similar tactics.
Ultimately, these findings underscore how viruses adapt in surprising ways to ensure their survival, even in unfamiliar hosts like humans. By reshaping human RNA to meet its needs, SARS-CoV-2 demonstrates a unique form of genetic flexibility, enabling it to bypass the limitations of its genetic makeup and thrive within human cells.
The study findings were published in the peer-reviewed International Journal of Molecular Sciences.
https://www.mdpi.com/1422-0067/25/21/11614
For the latest COVID-19 News, keep on logging to Thailand
Medical News.
Read Also:
https://www.thailandmedical.news/news/coronavirus-s-protein-alters-dsrna-accumulation-and-stress-granule-formation-by-regulating-adari1
https://www.thailandmedical.news/news/sars-cov-2-n-proteins-cause-degradation-of-cellular-components-involved-in-rna-interference-and-rna-splicing
