French Study Finds That Human Host Protein TDP-43 Plays A Role In Influenza A Virus Replication
Nikhil Prasad Fact checked by:Thailand Medical News Team Apr 30, 2024 11 months, 3 days, 3 hours, 43 minutes ago
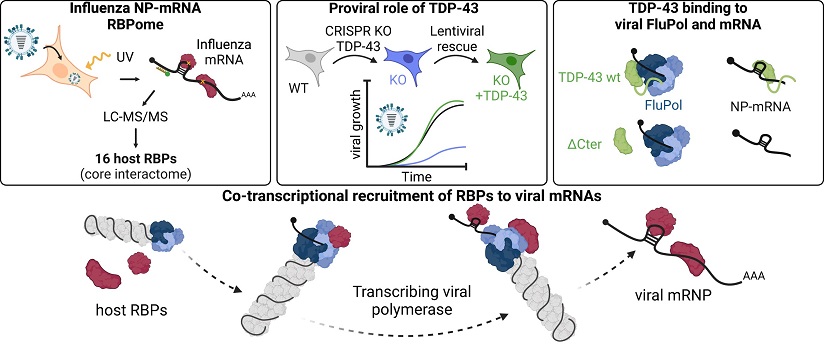
Influenza News: Influenza A viruses (IAVs) stand as significant pathogens, affecting both human and animal populations worldwide and causing substantial health and economic burdens. These viruses possess a genome comprising eight segments of negative-polarity viral RNA (vRNA), encoding ten major viral proteins and several auxiliary proteins. Central to viral replication is the formation of viral ribonucleoproteins (vRNPs), where vRNAs are encapsulated by nucleoprotein (NP) and viral RNA-dependent RNA polymerase (FluPol). These vRNPs undergo transcription, replication, and translation processes intricately intertwined with the host cellular machinery. Notably, the synthesis of viral mRNA involves a unique mechanism termed 'cap-snatching,' where the capped 5′ ends of host RNA polymerase II transcripts are cleaved by FluPol to initiate transcription, followed by polyadenylation and termination near the 5′ vRNA end.
 Graphical Abstract - Human Host Protein TDP-43 Plays A Role In Influenza A Virus Replication
Graphical Abstract - Human Host Protein TDP-43 Plays A Role In Influenza A Virus Replication
Despite significant progress in understanding RNA-binding proteins (RBPs) associated with cellular mRNAs, the RBPome of influenza virus mRNAs remains largely unexplored. This
Influenza News report delves into a recent study conducted by the Institut Pasteur, Université Paris Cité, CNRS-France, unraveling the role of the human host protein TDP-43 in influenza A virus replication.
Identifying Host Proteins Interacting with Influenza Virus NP-mRNA
Using an enhanced RNA interactome capture method, researchers identified 51 cellular proteins directly interacting with the NP-mRNA during influenza A virus infection. These proteins primarily belong to RNA/mRNA metabolic and splicing processes, with strong connections to nucleotide-binding domains and repeated RNA-binding domains characteristic of heterogeneous nuclear ribonucleoproteins (hnRNPs) and other RNA-binding proteins. Interestingly, the NP-mRNA interactome overlaps with cellular poly(A)-mRNA binders, suggesting shared protein compositions between viral and cellular mRNAs.
Among the identified proteins, TDP-43 emerged as a key player in influenza virus replication. Knock-out experiments demonstrated impaired viral replication in TDP-43-deficient cells, with subsequent rescue experiments restoring viral replication. Importantly, TDP-43's recruitment to viral mRNAs was mediated by the viral polymerase, primarily through protein-protein interactions rather than RNA-mediated binding. This recruitment mechanism highlights the intricate interplay between viral and host proteins in orchestrating viral mRNA processing and translation.
Exploring Biological Functions of Viral mRNA Binding Proteins
Gene Ontology (GO) enrichment analyses revealed that the NP-mRNA interactome is enriched in proteins involved in various mRNA processing steps, including transcription, splicing, transport, stability, and translation. Notably, sever
al proteins within this interactome have previously been implicated in influenza virus life cycle stages, reinforcing the dataset's validity.
Functional studies, including RNA interference screens targeting core interactome components, uncovered proteins crucial for viral infection, highlighting hnRNPA2B1, hnRNPH1, TDP-43, SF3B4, and SF3A3 as proviral factors promoting influenza virus replication.
Moreover, the study sheds light on the broader implications of RBP recruitment by viral polymerases, suggesting a conserved mechanism across divergent viruses and highlighting potential targets for antiviral therapies.
Role of TDP-43 in Viral Replication
The study provides insights into how the viral polymerase recruits RBPs like TDP-43 onto viral mRNAs, emphasizing a crucial role of the polymerase in assembling viral mRNPs. This recruitment not only affects viral mRNA stability and nuclear export but also hints at potential regulatory roles in mRNA translation. Moreover, the study sheds light on the broader implications of RBP recruitment by viral polymerases, suggesting a conserved mechanism across divergent viruses and highlighting potential targets for antiviral therapies.
The role of TDP-43 in viral replication represents a significant finding in the context of influenza A virus (IAV) infection. TDP-43, a well-known RNA-binding protein implicated in various cellular processes including mRNA processing and transport, emerges as a key player in the viral life cycle. The study delves into the intricate mechanisms through which TDP-43 influences viral replication, shedding light on crucial interactions between host RBPs and viral components.
-Recruitment Mechanisms
One of the pivotal discoveries is the mechanism of TDP-43 recruitment onto viral mRNAs by the viral polymerase. Unlike conventional RNA-mediated binding, TDP-43's association with viral mRNAs primarily occurs through protein-protein interactions facilitated by the viral polymerase. This unique recruitment mechanism underscores the adaptability of viral components in hijacking host cellular machinery to facilitate their own replication and gene expression. Understanding the nuances of this recruitment process opens avenues for targeted interventions aimed at disrupting viral-host protein interactions critical for viral replication.
-Impact on Viral mRNA Processing and Stability
TDP-43's presence on viral mRNAs influences various aspects of mRNA processing and stability. By binding to viral transcripts, TDP-43 potentially stabilizes these mRNAs within the nucleus, ensuring their efficient export to the cytoplasm for translation. Moreover, TDP-43's interaction with viral mRNAs may modulate their splicing patterns or influence the formation of viral ribonucleoprotein complexes (vRNPs), further impacting viral gene expression and replication efficiency. These insights into TDP-43's role in viral mRNA processing shed light on critical steps in the viral life cycle that can be targeted for therapeutic interventions.
-Nuclear Export and Translation
Another aspect highlighted is TDP-43's potential role in mediating nuclear export and subsequent translation of viral mRNAs. As viral mRNAs must be transported to the cytoplasm for efficient protein synthesis, TDP-43's presence on these transcripts could facilitate their export via host cellular export pathways. Additionally, TDP-43's interactions with components of the translational machinery may influence the efficiency and fidelity of viral protein synthesis, impacting overall viral replication dynamics. Unraveling the specifics of TDP-43's involvement in these processes provides crucial insights into viral-host interactions at the molecular level.
Implications for Antiviral Strategies
Understanding TDP-43's role in viral replication opens avenues for developing targeted antiviral strategies. By disrupting the recruitment of TDP-43 onto viral mRNAs or modulating its interactions with viral components, researchers can potentially inhibit viral gene expression and replication. Moreover, insights into TDP-43-mediated processes such as nuclear export and translation of viral mRNAs offer novel targets for antiviral drug development. These strategies, leveraging molecular insights into TDP-43's involvement in viral replication, hold promise for combating influenza A virus and other viral infections.
Future Directions
Further research is warranted to elucidate the precise mechanisms through which TDP-43 influences viral mRNA processing, stability, nuclear export, and translation. Investigating the dynamics of TDP-43-viral RNA interactions at different stages of infection and in diverse cellular contexts will deepen our understanding of viral-host interactions. Additionally, exploring the broader implications of TDP-43 recruitment by viral polymerases across different virus families could unveil common regulatory mechanisms with therapeutic potential beyond influenza A virus.
While this study significantly advances our understanding of influenza virus-host interactions, several avenues for future research emerge. Further investigations into the temporal and spatial dynamics of viral RNA interactomes, as well as the role of RBPs in viral mRNA processing and translation, are warranted.
Conclusion
The study underscores the intricate interplay between influenza A virus and host proteins, particularly highlighting the role of TDP-43 in viral replication. By unraveling the mechanisms of viral mRNA binding proteins and their recruitment by the viral polymerase, this research not only enhances our understanding of influenza virus-host interactions but also paves the way for developing targeted antiviral strategies leveraging these molecular insights.
The study findings were published in the peer reviewed journal: Nucleic Acids Research (Oxford Journals).
https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae291/7660082
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
