Glaucoma News: Breakthrough Discovery! Chinese Scientists Uncover Crucial circRNAs Implicated in Acute Angle-Closure Glaucoma!
Nikhil Prasad Fact checked by:Thailand Medical News Team Oct 10, 2023 1 year, 6 months, 1 week, 3 days, 19 hours, 49 minutes ago
Glaucoma News: Glaucoma, a group of degenerative diseases affecting the optic nerve, is the leading cause of irreversible blindness worldwide. With a prevalence of approximately 3.5% in individuals over the age of 40, glaucoma poses a significant global health challenge. Primary angle-closure glaucoma is especially prevalent among Asian populations, and its early diagnosis and treatment are essential to prevent vision loss. However, the lack of targeted biomarkers for glaucoma has hindered timely interventions.
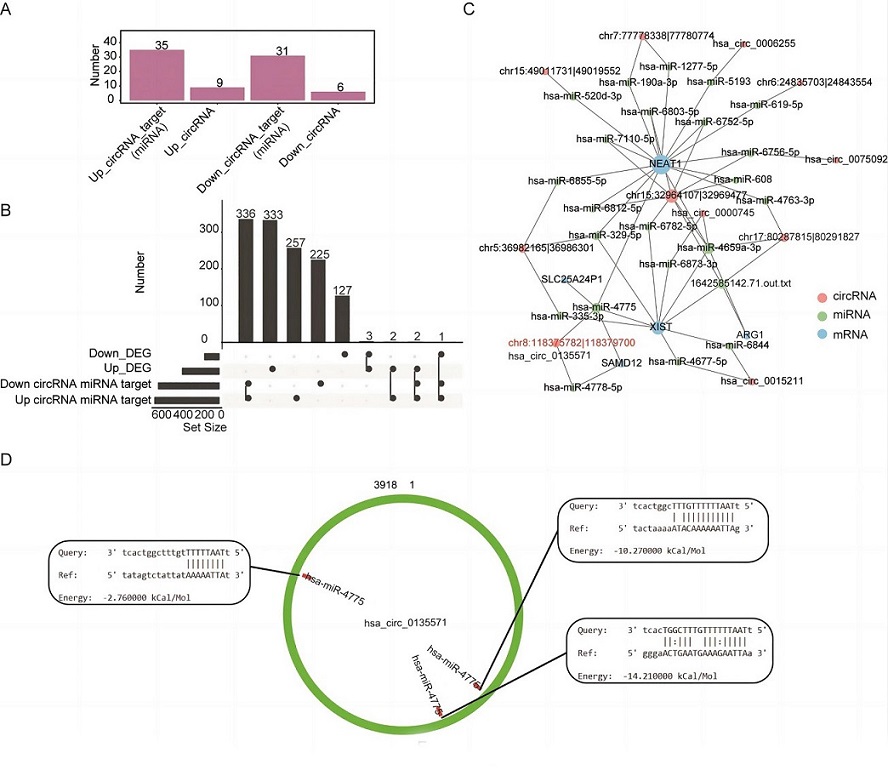 Construction of a circRNA–miRNA–mRNA network. (A) Number of miRNA targets of up- and downregulated differentially expressed circRNAs (DEcircRNAs) in the patient group. The miRNA targets of DEcircRNAs were obtained from the miRanda database. (B) Overlap analysis between the mRNA targets of miRNAs that are targeted by DEcircRNAs and differentially expressed genes (DEGs). The black dots indicate the gene groups of the overlap analysis. The bars with numbers indicate the number of genes of each group or overlapped group. (C) Network of DEGs, DEcircRNAs, and miRNAs. (D) Binding site of hsa-miR-4475 on circRNA hsa-circ_0135571.
Construction of a circRNA–miRNA–mRNA network. (A) Number of miRNA targets of up- and downregulated differentially expressed circRNAs (DEcircRNAs) in the patient group. The miRNA targets of DEcircRNAs were obtained from the miRanda database. (B) Overlap analysis between the mRNA targets of miRNAs that are targeted by DEcircRNAs and differentially expressed genes (DEGs). The black dots indicate the gene groups of the overlap analysis. The bars with numbers indicate the number of genes of each group or overlapped group. (C) Network of DEGs, DEcircRNAs, and miRNAs. (D) Binding site of hsa-miR-4475 on circRNA hsa-circ_0135571.
Circular RNAs (circRNAs), a class of noncoding RNAs, have emerged as key players in various biological processes. They act as microRNA (miRNA) sponges and are implicated in the regulation of gene expression. Consequently, circRNAs have garnered attention as potential biomarkers and therapeutic targets in numerous human diseases. Despite this, the precise mechanisms by which circRNAs contribute to the pathogenesis of glaucoma have remained elusive.
In a groundbreaking study conducted by researchers from Qinghai Provincial People’s Hospital in China and the China Academy of Chinese Medical Sciences in Beijing, transcriptome sequencing was employed to identify differentially expressed circRNAs (DEcircRNAs) in peripheral blood samples from patients with primary angle-closure glaucoma. The research covered in this
Glaucoma News report aimed to shed light on the potential roles of DEcircRNAs in glaucoma and explore their utility as biomarkers and therapeutic targets.
Transcriptome Sequencing Analysis
The transcriptome sequencing analysis of blood samples collected from glaucoma patients revealed notable differences in gene expression profiles between the patient and control groups. A total of 481 differentially expressed genes (DEGs) were identified, with 350 genes upregulated and 131 genes downregulated. Functional analysis using Gene Ontology (GO) revealed enrichment in pathways related to complement activation, immune response, and small molecule metabolic processes among upregulated DEGs. Conversely, downregulated DEGs were associated with extracellular matrix organization, innate immune response, and signal transduction.
Furthermore, several genes, including CFD, IGHG1, IGHA1, and noncoding RNAs like SNORD3A, SNORD16, SNORA52, and SNORA23, exhibited differential expression in glaucoma patients, further underscoring their potential relevance to the disease.
Identification and Characteristics of Glaucoma-Related circRNAs
The study team also conducted circRNA sequencing analysis and identified 345 DEcircRNAs in peripheral blood samples from glaucoma patients, with 278 circRNAs upregulated and 67 circRNAs downregulated. Bioinformatics analysis was performed to determine the host genes of these DEcircRNAs, and GO analysis was used to elucidate their functional roles.
The results demonstrated that upregulated DEcircRNAs were associated with processes such as mitotic cell cycle, regulation of protein localization, and cellular lipid metabolism, while downregulated DEcircRNAs were enriched in terms like DNA-dependent transcriptional regulation. Intriguingly, seven host genes (NEAT1, SAMD12, KDM5D, ZFY, EIF1AY, TXLNGY, and DDX3Y) of nine DEcircRNAs were also identified among the DEGs, highlighting the consistency of their expression patterns.
Validation of Glaucoma-Associated DEGs
The DEGs presumed to be associated with glaucoma were further validated through quantitative reverse transcription PCR (qRT-PCR) analysis. This validation confirmed the significant upregulation of genes such as IGHG1, CFD, IGHA1, TMEM107, SNHG3, OLIG1, SNORD3A, SNORD16, SNORA52, and SNORA23 in glaucoma patients, along with the downregulation of CLEC12A, consistent with the findings from RNA sequencing.
.jpg) Co-Expression Network of DEcircRNAs and DEGs in Glaucoma
Co-Expression Network of DEcircRNAs and DEGs in Glaucoma
CircRNAs have been shown to act as miRNA sponges, regulating the expression of target genes. In this study, potential binding pairs of miRNAs and DEcircRNAs were identified from the miRanda database, suggesting that DEcircRNAs may contribute to the differential expression of DEGs associated with glaucoma.
The analysis revealed that nine upregulated and six downregulated DEcircRNAs had the potential to bind to 35 and 31 miRNAs, respectively. These DEcircRNAs and miRNAs could form 67 interacting pairs, as indicated by miRanda scores. Furthermore, these 64 miRNAs had the potential to target 823 genes. Overlapping analysis of the 823 genes with the 481 DEGs identified five DEGs as potential targets of these miRNAs.
A comprehensive binding-target network was constructed, illustrating how 11 DEcircRNAs might regulate the expression of five DEGs (NEAT1, XIST, ARG1, SAMD12, and SLC25A24P1) through interactions with 23 miRNAs. Notably, NEAT1 emerged as a primary target of a substantial proportion of these miRNAs and DEcircRNAs, with hsa_circ_0135571 being implicated in the regulation of NEAT1 expression by binding to hsa-miR-4775.
Validation of circRNA Expression Patterns
To validate the expression patterns, hsa_circ_0135571 and NEAT1 were subjected to RT-qPCR analysis in blood samples from glaucoma patients and healthy controls. The results demonstrated significant upregulation of both hsa_circ_0135571 and NEAT1 in glaucoma patients compared to the control group, providing additional evidence for their involvement in glaucoma pathogenesis.
Conclusion
This groundbreaking study has unveiled critical insights into the role of circRNAs in the context of glaucoma, particularly in primary angle-closure glaucoma. The identification of DEcircRNAs and their potential regulatory functions through miRNAs has significant implications for understanding the molecular mechanisms underlying glaucoma development.
One circRNA, hsa_circ_0000745, was highlighted as a potential biomarker for early glaucoma diagnosis, with its regulation of NEAT1 expression through miRNA interactions. Nevertheless, it's important to note that further validation and large-scale studies are necessary to confirm the reliability and reproducibility of circRNAs as biomarkers for diagnosing and treating glaucoma.
In conclusion, the research underscores the importance of circRNAs in glaucoma pathogenesis, offering a glimmer of hope for early diagnosis and targeted therapies that could potentially transform the landscape of glaucoma management.
The study findings were published in the peer reviewed journal: Scientific Reports. (Nature).
https://www.nature.com/articles/s41598-023-44073-y
For the latest
Glaucoma News, keep on logging to Thailand Medical News.

.jpg)