Indian Study Reveals That Of 2896 Human Host Proteins That Interact With 29 SARS-CoV-2 Proteins, 176 Host Genes Interacts With All Viral Proteins!
Nikhil Prasad Fact checked by:Thailand Medical News Team Apr 20, 2024 1 year, 6 days, 12 hours, 38 minutes ago
COVID-19 News: The COVID-19 pandemic has sparked an unprecedented wave of scientific inquiry into the intricacies of the SARS-CoV-2 virus and its interactions with human host cells. Among the myriad research endeavors, a collaborative effort that is covered in this
COVID-19 News report, involving esteemed institutions such as the All India Institute of Medical Sciences (AIIMS) in Delhi, the Indian Institute of Technology (IIT) in Hauz Khas, New Delhi, King Abdulaziz University in Saudi Arabia, University of Jeddah in Saudi Arabia, Jawaharlal Nehru University in New Delhi, India, and the University of Pittsburgh Medical Center in the USA has yielded groundbreaking insights into the molecular landscape of COVID-19.
 Of 2896 Human Host Proteins That Interact With 29 SARS-CoV-2 Proteins, 176 Host Genes Interacts With All Viral Proteins
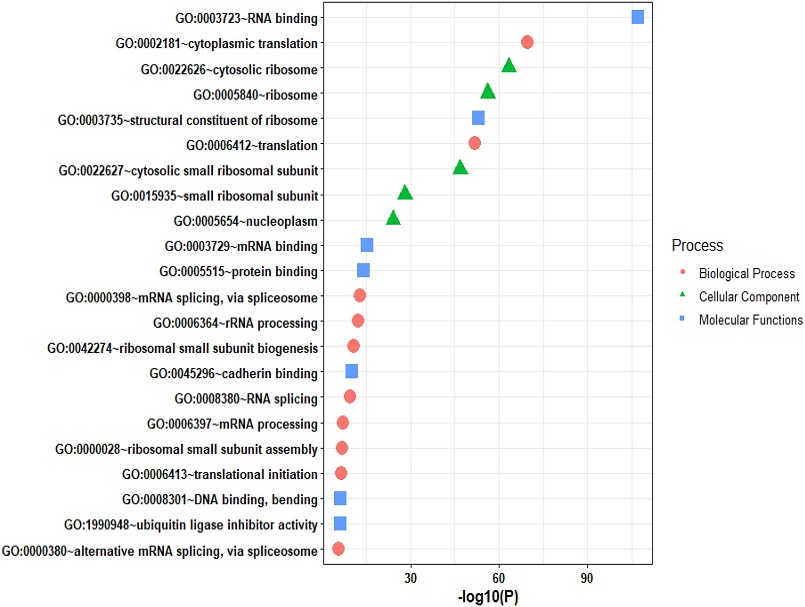
Gene ontology analysis of the identified 176 host proteins showing direct interaction with viral proteins
Study Design and Methodology
Of 2896 Human Host Proteins That Interact With 29 SARS-CoV-2 Proteins, 176 Host Genes Interacts With All Viral Proteins
Gene ontology analysis of the identified 176 host proteins showing direct interaction with viral proteins
Study Design and Methodology
Employing a sophisticated network-based approach, the study meticulously constructed a complex network comprising 29 SARS-CoV-2 proteins and their interactions with 2896 human host proteins. This analytical framework aimed to unravel the intricate web of molecular interactions underpinning viral infection pathways. Notably, the study identified 176 host genes that exhibited interactions with all 29 viral proteins, indicative of their pivotal roles in the virus-host interplay.
Exploring Biological Processes and Pathways
Gene ontology and pathway analyses were instrumental in elucidating the biological processes in which these host proteins are intricately involved. Key processes highlighted by the analyses included translation, mRNA splicing, ribosomal pathways, and cytoplasmic translation. These findings underscored the multifaceted nature of host-virus interactions and provided crucial insights into the molecular mechanisms driving COVID-19 pathogenesis.
Identification of Hub Genes
Within the intricate network of protein-protein interactions (PPI), five hub genes emerged as central players: EEF2, RPS3, RPL9, RPS16, and RPL11. These hub genes exhibited significant connectivity within the disease-associated network, indicating their critical roles in orchestrating molecular responses during viral infections. Further analysis revealed that these hub genes are primarily involved in ribosomal pathways, cytoplasmic translation, and mRNA splicing, underscoring their importance in COVID-19 pathogenesis.
Insights into Disease-Gene Interactions
A disease-gene interaction network was constructed to shed light on the comorbidities associated with COVID-19 patients. This comprehensive analysis revealed associations between the hub genes and various disorders such as respiratory dif
ficulties, dysarthria, congestive heart failure, melanoma, anaemia, Parkinson's disease, cerebellar atrophy, and truncal ataxia. The network highlighted shared genetic factors among different disorders, providing valuable insights into the complex interplay between COVID-19 and other health conditions.
Exploring Drug Repurposing Opportunities
One of the study's significant contributions was the exploration of drug repurposing opportunities based on interactions with hub genes. A thorough screening of drug-gene interaction databases, including PubChem, DrugBank, and CTD, identified 292 drug molecules with interactions with hub genes. Among these, 13 molecules exhibited interactions with all five hub genes, pointing towards potential therapeutic candidates for COVID-19. Notably, estradiol, a steroid hormone, emerged as a top candidate due to its interactions with all hub genes involved in protein synthesis and immune response.
Potential Therapeutic Implications of Estradiol
The study's analysis revealed intriguing insights into the therapeutic potential of estradiol in combating COVID-19. Estradiol, known for its role in regulating various physiological processes, demonstrated interactions with key hub genes critical for protein synthesis and immune response. Further analysis using the Enrichr database and Gene Expression Omnibus (GEO) profiles highlighted the potential of estradiol to upregulate the expression of hub genes, thereby modulating essential molecular pathways implicated in COVID-19 pathogenesis.
Implications for Future Research and Clinical Translation
The comprehensive findings of this study have significant implications for future research and clinical translation. Firstly, the identification of hub genes and their roles in COVID-19 pathogenesis provides a robust foundation for targeted therapeutic interventions. Secondly, the exploration of drug repurposing avenues underscores the importance of leveraging existing pharmacological agents to combat emerging viral threats. Thirdly, the potential therapeutic value of estradiol opens new avenues for the development of hormone-based therapies for COVID-19.
Conclusion
In conclusion, the collaborative effort between leading institutions has yielded invaluable insights into the molecular landscape of COVID-19. The identification of hub genes, exploration of disease-gene interactions, and elucidation of drug repurposing opportunities represent significant strides towards understanding and combating the pandemic. The potential therapeutic implications of estradiol underscore the importance of innovative approaches in the quest for effective COVID-19 treatments. Moving forward, continued research and clinical validation are imperative to translate these findings into tangible therapeutic interventions and mitigate the global impact of the pandemic.
The study findings were published in the peer reviewed journal: Heliyon.
https://www.sciencedirect.com/science/article/pii/S240584402405998X
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
