Monash University Scientists Uncover A Novel Fold in SARS-CoV-2 Replication Co-Factor Non-Structural Protein 9 - A New Potential Therapeutic Target
COVID-19 News -Novel Fold - Nsp9 Apr 16, 2023 2 years, 4 days, 15 hours, 11 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had a devastating impact on global health and the economy. The development of vaccines and therapeutic agents targeting various viral components is crucial to combat the virus effectively. In a recent study by researchers from Monash University-Australia, the study team uncovered a novel fold in a key protein involved in SARS-CoV-2 replication, known as non-structural protein 9 (nsp9).
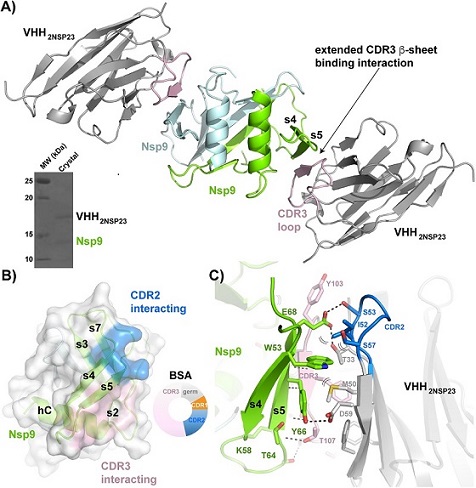 VHH2nsp23-bound SARS-CoV-2 nsp9cov19. A) Cartoon representation of the two VHH2nsp23: nsp9COV19 complexes within the asymmetric unit of the crystals. The two VHH2nsp23 subunits are shown in grey with the extending VHHCDR3β-sheet interacting residues in pink. The two nsp9COV19 subunits are coloured green and light blue. The insert shows the intact complex run on SDS-PAGE after data collection. B) An overlayed surface representation of nsp9COV19 coloured according to the regions bound by the VHH—VHHCDR2 interacting residues are highlighted with blue colouring and the VHHCDR3 residues with pink colouring. The doughnut chart insert shows the respective buried surface area percentages. C) An enhanced view of the residue interactions across the VHH2nsp23: nsp9COV19 interface. Hydrogen bonds are denoted with dashed lines and van der Waals interactions with double brackets.
Nsp9's Role in SARS-CoV-2 Replication
VHH2nsp23-bound SARS-CoV-2 nsp9cov19. A) Cartoon representation of the two VHH2nsp23: nsp9COV19 complexes within the asymmetric unit of the crystals. The two VHH2nsp23 subunits are shown in grey with the extending VHHCDR3β-sheet interacting residues in pink. The two nsp9COV19 subunits are coloured green and light blue. The insert shows the intact complex run on SDS-PAGE after data collection. B) An overlayed surface representation of nsp9COV19 coloured according to the regions bound by the VHH—VHHCDR2 interacting residues are highlighted with blue colouring and the VHHCDR3 residues with pink colouring. The doughnut chart insert shows the respective buried surface area percentages. C) An enhanced view of the residue interactions across the VHH2nsp23: nsp9COV19 interface. Hydrogen bonds are denoted with dashed lines and van der Waals interactions with double brackets.
Nsp9's Role in SARS-CoV-2 Replication
Nsp9 is a small accessory factor that plays an essential role in coronavirus replication and the replication transcription complex (RTC) as covered in previous studies and
COVID-19 News coverages. It associates with the N-terminal pseudokinase Nidovirales ribonucleic acid (RNA)-dependent RNA polymerase (RdRp) associated nucleotidyltransferase domain (NiRAN) of nsp12, which is produced inside host cells as a self-cleaving PP1ab polyprotein. Nsp12 contains a vital viral RdRp, which, along with nsp7 and nsp8, becomes the core component of RTC.
Nsp9 is characterized by its distinct viral fold, RNA-binding protein properties, and being a key element for viral messenger RNA (mRNA) capping. It also recruits other proteins for viral 5’-mRNA capping, an essential aspect of viral replication.
The nsp9 and NiRAN domain act jointly as a polyribonucleotidyltransferase (PRNTase) with catalytic and adduct-accepting residues on different amino acid chains, making nsp12 a potentially viable therapeutic target.
Nanobodies and Their Potential as Therapeutics
Camelid immunoglobulins (Ig)-derived nanobodies, also known as variable heavy domains (VHHs), have demonstrated high specificity for nsp9. Several studies have identified small molecules with affinity for nsp9COV19 that could potentially inhibit NiRAN engagement, preventing SARS-CoV-2 RNAylation and capping.
Nanobodies, as specific reagents, could aid in understanding nsp9's PRNTase role and could represent starting points for broad antivirals.
The Study: Investigating the Flexibility of Nsp9
In the present study, the study team aimed to des
cribe the flexibility of nsp9. To do this, they purified nsp9 and the anti-nsp9 VHH2nsp23 complex and then analyzed the crystal structure of the complex at a resolution of 2.4Å using X-ray diffraction. The team built and refined the crystal structure of the VHH2nsp23-bound state of nsp9COV19, which showed two copies of the nsp9COV19: VHH2nsp23 complex within the asymmetric unit overlay.
The study team found transformation-related protein 53 (Trp-53) to be a primary feature of the extensive antibody-binding interface of nsp9COV19, with the CDR3-loop forming an extended β-sheet interaction. Nanobody binding triggered large-scale topological changes to the unique coronaviral fold of nsp9, distorting all NiRAN-interacting elements of nsp9.
The researchers explored potential points of flexibility within the nsp9 fold and compared bound and unbound states of nsp9COV19 using a Kleywegt plot and mapping Ramachandran distances between each state onto the nsp9COV19 structure.
Key Findings
The study confirmed that the binding mode of anti-nsp9 specific nanobodies is centered upon Trp-53 within SARS-CoV-2 nsp9. Antibody binding at this site surprisingly results in large-scale changes to the overall topology of this coronaviral unique fold. The study team further characterized the antibody-induced structural dynamism within nsp9, identifying a number of potentially flexible regions. A large expansion of the cavity between the s2-s3 and s4-s5 loops is particularly noteworthy. As is the potential for large-scale movements in the C-terminal GxxxG helix.
The study revealed that the VHH 2nsp23-induced conformational changes in nsp9 disrupt the NiRAN-binding surface, which is essential for viral replication. The study team discovered a new loop-helix-loop motif in nsp9COV19 and identified it as a potential target for future therapeutic interventions. Additionally, they characterized the flexibility of nsp9 and its potential to adapt to different binding partners.
The study findings also highlight the potential of nanobodies as both tools for studying protein conformational changes and as starting points for the development of antiviral therapies.
The unique fold of nsp9 and its role in viral replication make it an attractive target for therapeutics, and the study's results provide valuable structural insights that can guide the design of small molecules or biologics to inhibit nsp9 function.
The study team also noted that the VHH2nsp23-bound structure appears to allow more freedom for the protein’s N-terminus following loss of the Tyr-cradle. The study findings highlight a number of structural points within the nsp9 fold that may be capable of acting as hinge points. Moreover, the study findings demonstrated that VHH-binding, which occurs at a site distal to the NiRAN domain-interaction site, may nonetheless result in structural changes capable of disrupting a protein-interface. The potential for alternate forms of nsp9, as well as issues over its oligomerization state, should to some extent be kept in mind when assessing in vivo mutational experiments.
Conclusion
This study sheds light on the flexibility of nsp9 and uncovers a novel fold in the protein, providing essential structural insights into its function. The identification of this new motif and the conformational changes induced by nanobody binding offer potential avenues for the development of novel therapeutics targeting nsp9. The use of nanobodies as specific reagents can not only help in understanding the protein's PRNTase role but also provide a starting point for the development of broad antiviral agents against SARS-CoV-2 and potentially other coronaviruses.
The study findings were published in the peer reviewed journal: PLOS One.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0283194
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
