Nikhil Prasad Fact checked by:Thailand Medical News Team Jul 15, 2024 1 year, 7 months, 1 week, 6 days, 14 hours, 23 minutes ago
COVID-19 News: Understanding the Mutations
Scientists from various renowned institutions, including the Institut de Biologia Molecular de Barcelona-Spain and the Centro Nacional de Biotecnología in Madrid-Spain, have made significant strides in understanding how specific mutations in the SARS-CoV-2 virus affect its ability to replicate. This
COVID-19 News report explores their groundbreaking findings, highlighting the potential implications for future treatments and vaccines.
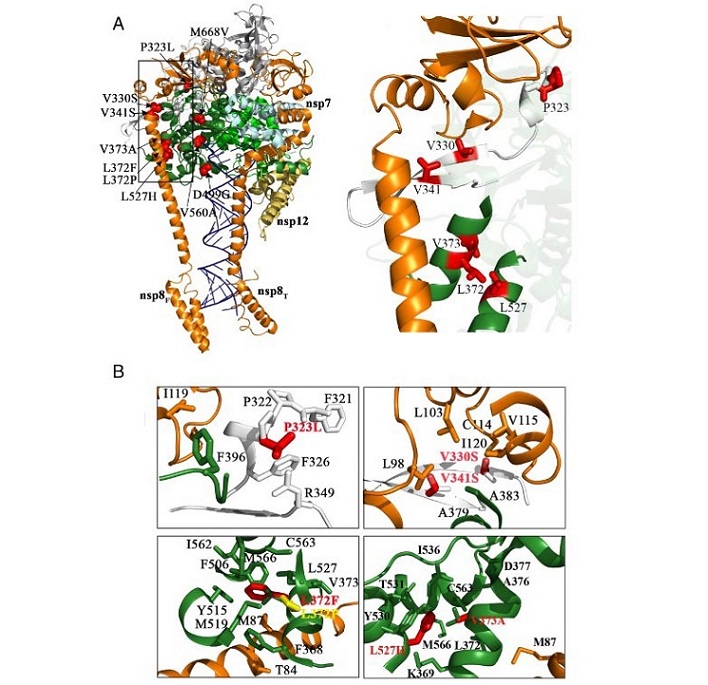 Location and functional implication of the selected amino acid substitutions in the nsp12 protein of SARS-CoV- 2. (A) The Left panel shows the location of the amino acid substitutions studied here (P323L, V330S, V341S, L372F, L372P, V373A, D499G, L527H, V560A, and M668V) in the three-dimensional structure of nsp12. The structure used as reference is that of the nsp12–nsp8–nsp7–RNA complex (PDB: 6YYT), depicted as a cartoon representation with nsp12 colored by domains (NiRAN in gray, interface in white, and the RdRp domain in dark green, green, and dark yellow for the fingers, palm, and thumb subdomains, respectively). The cofactors nsp8 and nsp7 are colored in orange and cyan, respectively, and the RNA is depicted in dark blue. Substituted amino acids are shown as red spheres and explicitly labeled. The Right panel shows a close-up view, highlighting the positions of the amino acid substitutions in the nsp12–nsp8F contact interface (side chains in red sticks). (B) Detailed views of the interactions around the mutated positions. Side chains of substituted amino acids and neighboring residues are shown as sticks in different colors (white for residues in the nsp12 interface domain, green for residues in the nsp12 fingers, and orange for nsp8), and explicitly labeled. Both wild type and mutated side chains are seen in the different panels. The Upper Left panel shows the P323L substitution placed in thensp12 interface (depicted in white for proline and in red for the mutated leucine). The Upper Right panel shows substitutions V330S and V341S, within the nsp12interface (white for the valine side chains and red for the mutated serine residues). The Bottom Left panel shows L372F and L372P side chain substitutions, within de nsp12 fingers (side chain depicted in green for leucine, red for phenylalanine, and yellow for proline). The Bottom Right panel shows V373A and L527Hsubstitutions, also within the nsp12 fingers (green for valine and leucine side chains and red for alanine and histidine).
The Study and Its Importance
Location and functional implication of the selected amino acid substitutions in the nsp12 protein of SARS-CoV- 2. (A) The Left panel shows the location of the amino acid substitutions studied here (P323L, V330S, V341S, L372F, L372P, V373A, D499G, L527H, V560A, and M668V) in the three-dimensional structure of nsp12. The structure used as reference is that of the nsp12–nsp8–nsp7–RNA complex (PDB: 6YYT), depicted as a cartoon representation with nsp12 colored by domains (NiRAN in gray, interface in white, and the RdRp domain in dark green, green, and dark yellow for the fingers, palm, and thumb subdomains, respectively). The cofactors nsp8 and nsp7 are colored in orange and cyan, respectively, and the RNA is depicted in dark blue. Substituted amino acids are shown as red spheres and explicitly labeled. The Right panel shows a close-up view, highlighting the positions of the amino acid substitutions in the nsp12–nsp8F contact interface (side chains in red sticks). (B) Detailed views of the interactions around the mutated positions. Side chains of substituted amino acids and neighboring residues are shown as sticks in different colors (white for residues in the nsp12 interface domain, green for residues in the nsp12 fingers, and orange for nsp8), and explicitly labeled. Both wild type and mutated side chains are seen in the different panels. The Upper Left panel shows the P323L substitution placed in thensp12 interface (depicted in white for proline and in red for the mutated leucine). The Upper Right panel shows substitutions V330S and V341S, within the nsp12interface (white for the valine side chains and red for the mutated serine residues). The Bottom Left panel shows L372F and L372P side chain substitutions, within de nsp12 fingers (side chain depicted in green for leucine, red for phenylalanine, and yellow for proline). The Bottom Right panel shows V373A and L527Hsubstitutions, also within the nsp12 fingers (green for valine and leucine side chains and red for alanine and histidine).
The Study and Its Importance
In this recent study, researchers focused on the RNA-dependent RNA polymerase (RdRp), a crucial enzyme in the virus responsible for replicating its RNA. By examining diagnostic samples from patients during the first wave of the COVID-19 pandemic, they identified 41 amino acid substitutions in the RdRp protein nsp12. The team then selected eight of these substitutions to investigate their impact on the replication complex, which includes the nsp12, nsp8, and nsp7 proteins.
Key Findings
The researchers discovered that mutations near the nsp12-nsp8 interface, specifically in a hydrophobic cluster within the nsp12 fingers subdomain, significantly affect the virus's RNA polymerization activity. This cluster, involving contacts between helices in nsp12 and the long alpha-helix of nsp8, plays a critical role in the modulation of RdRp activity.
Three mutations (D499G, M668V, and V560A) around the polymerase central cavity showed similar polymerization rates to the wild type (WT) RdRp. However, five other mutations (P323L, L372F, L372P, V373A, and L527H) near the nsp12-nsp8 contact surface resulted in significant changes in RNA polymerization activity.
Detailed Analysis
The study highlighted that the mutation P323L, which has persisted since early 2020, showed enhanced RdRp activity at both 33°C and 37°C compared to the WT RdRp. This mutation, first observed in Europe, was associated with a higher mutation rate and better binding to the antiviral drug Remdesivir.
Mutations L372F, L372P, V373A, and L527H, found at low frequencies in patient samples, displayed varying effects on RdRp activity. For instance, the L372F mutation increased polymerization activity, while L372P, L527H, and V373A weakened the hydrophobic interactions, reducing RNA synthesis efficiency. These mutations disrupt the hydrophobic clusters, crucial for stabilizing the nsp12-nsp8 interaction, underscoring their importance in regulating viral replication.
Additional Insights
The study further examined mutations designed to disrupt the nsp12-nsp8 interactions, such as nsp12-V330S, nsp12-V341S, and nsp8-R111A/D112A. These mutations also resulted in impaired RdRp activity, reinforcing the importance of these contact surfaces in RNA synthesis.
The researchers used various biochemical assays to measure the RNA synthesis activities of the mutated RdRp complexes. Primer extension assays showed that while some mutations enhanced activity, others significantly reduced it.
Additionally, Electrophoretic Mobility Shift Assays (EMSA) revealed differences in RNA-binding capacities among the mutants, providing further insights into how these mutations affect the viral replication machinery.
Implications for Treatment and Vaccines
Understanding how these mutations impact the virus's replication can help in developing more effective antiviral drugs and vaccines. Targeting the critical nsp12-nsp8 interaction interface might offer new avenues for therapeutic intervention. As the virus continues to evolve, this knowledge is crucial for anticipating potential changes that could affect treatment efficacy.
Conclusion
This study findings offers valuable insights into the genetic variability of SARS-CoV-2 and its implications for viral replication and treatment strategies.
The findings were published in the peer-reviewed journal PNAS, highlighting the importance of ongoing research in combating the COVID-19 pandemic.
https://www.pnas.org/doi/10.1073/pnas.2317977121
For the latest
COVID-19 News, keep on logging to
Thailand Medical News.
Read Also:
https://www.thailandmedical.news/news/under-immune-pressure-sars-cov-2-gives-rise-to-e484d-and-l1265r-h1271y-mutations
https://www.thailandmedical.news/news/hyperactive-mutations-in-sars-cov-2-mpro-contribute-to-antiviral-drug-resistance
