Nikhil Prasad Fact checked by:Thailand Medical News Team Oct 24, 2024 5 months, 2 weeks, 5 days, 20 hours, 35 minutes ago
Medical News: A Closer Look at Reticulophagy
Reticulophagy is an essential cellular process that helps maintain the balance within cells by degrading parts of the endoplasmic reticulum (ER). This intricate system ensures that cells can respond to various types of stress and maintain proper functioning. Now, a recent study has shed light on the role of reticulophagy in viral infections, suggesting that this process may be more crucial than previously understood.
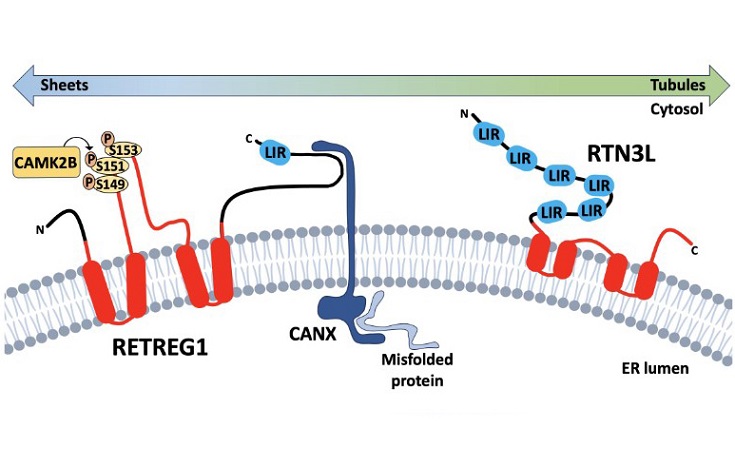 Reticulon homology domain-containing reticulophagy receptors RETREG1 and RTN3L. Reticulon homology domain (rhd)-containing reticulophagy receptorsare comprised of transmembrane or membrane-associated RHD domains (in red) connected to domains involved in substrate recognition and LC3 binding. RETREG1is primarily found in ER sheets where it captures misfolded substrate proteins bound to the ER transmembrane chaperone CANX. ER stress triggers post-translationalmodifications of the RETREG1 RHD, including CAMK2B-mediated phosphorylation, that contribute to RETREG1 oligomerization and er-scission, while LC3 bindingrecruits phagophores to initiate local degradation of ER membranes and cargo. RTN3L is primarily associated with the cytoplasmic leaflet of ER tubules, and relativelylittle is known about substrate recognition mechanisms. Six LIRs on the cytoplasmic amino-terminal domain direct binding to Atg8-like proteins and phagophorerecruitment.
Reticulon homology domain-containing reticulophagy receptors RETREG1 and RTN3L. Reticulon homology domain (rhd)-containing reticulophagy receptorsare comprised of transmembrane or membrane-associated RHD domains (in red) connected to domains involved in substrate recognition and LC3 binding. RETREG1is primarily found in ER sheets where it captures misfolded substrate proteins bound to the ER transmembrane chaperone CANX. ER stress triggers post-translationalmodifications of the RETREG1 RHD, including CAMK2B-mediated phosphorylation, that contribute to RETREG1 oligomerization and er-scission, while LC3 bindingrecruits phagophores to initiate local degradation of ER membranes and cargo. RTN3L is primarily associated with the cytoplasmic leaflet of ER tubules, and relativelylittle is known about substrate recognition mechanisms. Six LIRs on the cytoplasmic amino-terminal domain direct binding to Atg8-like proteins and phagophorerecruitment.
Researchers from the Department of Microbiology and Immunology at Dalhousie University in Canada, Alexa Wilson and Craig McCormick, have reviewed the latest evidence linking reticulophagy to the mechanisms viruses use to interact with and subvert the host's cellular machinery. Their findings, published in the peer-reviewed journal Autophagy, suggest that this cellular process plays both protective and detrimental roles in viral infections, depending on the context.
What Is Reticulophagy?
Reticulophagy is a specialized form of autophagy, a process where a cell degrades and recycles its components. Autophagy typically involves the degradation of cellular structures like mitochondria, but reticulophagy specifically targets the ER for degradation.
The ER is a large and complex cellular structure where proteins and lipids are synthesized and processed. When cells are under stress, such as during viral infection, the ER can become overloaded with misfolded proteins. Reticulophagy helps by breaking down parts of the ER that are no longer needed or that are malfunctioning.
This
Medical News report highlights that viruses have evolved ways to manipulate reticulophagy to create more favorable conditions for their replication. This manipulation can include evading immune responses or creating viral factories within the ER, where they can reproduce without detection.
How Do Viruses Use Reticulophagy?
The authors of the study identify several ways in which viruses use reticulophagy to their advantage. For example, many viruses create structures within the ER called double-membrane vesicles (DMVs) that serve as sites for viral replication. These DMVs shield the
virus from the host’s immune system and help concentrate the materials needed for viral reproduction.
Some viruses, such as coronaviruses (including SARS-CoV-2), are known to manipulate reticulophagy receptors like RETREG1 to inhibit the process, allowing the virus to avoid being degraded by the cell. By inhibiting reticulophagy, viruses prevent the cell from eliminating infected parts of the ER and allow viral replication to proceed unchecked.
Reticulophagy as an Antiviral Defense
However, reticulophagy is not always on the virus's side. This process can also serve as a form of antiviral defense by degrading viral components and limiting their replication. In some cases, reticulophagy can help clear viral proteins from the ER and prevent the virus from creating a hospitable environment for replication.
The study refers to this defense as "xERophagy," a term coined by the researchers to describe the specific degradation of viral components via reticulophagy. The authors reviewed multiple examples where reticulophagy appears to slow down or limit viral infections, such as with flaviviruses like Dengue and Zika viruses.
Key Players in Reticulophagy
Several receptors and proteins play crucial roles in reticulophagy. The discovery of the first mammalian reticulophagy receptor, RETREG1 (also known as FAM134B), was a breakthrough in the field. RETREG1 helps to identify parts of the ER that need to be degraded and recruits other cellular components to assist in the degradation process.
Other important proteins in this process include calnexin, which binds to misfolded proteins and helps target them for degradation, and reticulons like RTN3, which are involved in reshaping the ER during reticulophagy.
Viruses often target these proteins to subvert the reticulophagy process. For example, flaviviruses produce proteases that cleave RETREG1, preventing it from degrading the ER components that the virus needs to replicate.
How Does Reticulophagy Impact Different Viruses?
The study provides several examples of how different viruses interact with the reticulophagy process. For example, flaviviruses like Zika and Dengue viruses rely on the ER to create replication compartments. These viruses suppress reticulophagy to ensure that their replication compartments are not degraded.
On the other hand, viruses like Ebola have been shown to be limited by reticulophagy. In cells that are deficient in reticulophagy receptors like RETREG1, Ebola virus replication increases, suggesting that the degradation of ER components is a crucial part of the body’s defense against this virus.
The study also highlights the role of reticulophagy in coronavirus infections. Both SARS-CoV-1 and SARS-CoV-2 produce proteins that inhibit reticulophagy, allowing the virus to replicate more efficiently within the ER. However, the exact mechanisms by which these proteins interfere with reticulophagy are still being investigated.
Implications for Future Research
Understanding the dual role of reticulophagy in viral infections opens up new avenues for research and potential treatments. By enhancing the antiviral functions of reticulophagy or preventing viruses from inhibiting the process, it may be possible to develop new therapies that limit viral replication.
For example, drugs that target viral proteases responsible for cleaving RETREG1 could potentially restore the cell’s ability to degrade viral components. Similarly, enhancing the cell’s natural reticulophagy processes could help prevent the creation of viral replication compartments.
The study emphasizes the need for further research into the specific mechanisms viruses use to manipulate reticulophagy and how these processes can be countered by the host’s immune system.
Conclusion
Reticulophagy is an essential process that helps maintain cellular health by degrading and recycling parts of the ER. While viruses have evolved ways to manipulate this process to their advantage, reticulophagy also serves as a form of antiviral defense.
This study provides new insights into how reticulophagy interacts with viral infections and highlights the potential for future research to develop new antiviral therapies based on this process. By better understanding the mechanisms at play, researchers may be able to develop drugs that enhance the body’s natural defenses or prevent viruses from hijacking reticulophagy.
The study findings were published in the peer-reviewed journal: Autophagy.
https://www.tandfonline.com/doi/full/10.1080/15548627.2024.2414424
For the latest on Reticulophagy, keep on logging into Thailand
Medical News.
Read Also:
https://www.thailandmedical.news/news/russian-study-finds-that-myelin-basic-protein-antagonizes-sars-cov-2-orf3a-induced-autophagy-inhibition
https://www.thailandmedical.news/news/unraveling-the-intricacies-of-sars-cov-2-evolution-the-role-of-envelope-protein-mutation-t9i-in-autophagy-resistance
https://www.thailandmedical.news/news/sars-cov-2-causes-downregulation-of-autophagy-genes-pik3c3-and-rab7a
