New Study Uncovers Similarities and Differences Between Similarities Between SARS-CoV-2 And HIV-1 Infections In Terms Of Cellular And Immune Responses
Source: SARS-CoV-2-HIV Jan 15, 2022 3 years, 3 months, 3 days, 5 hours, 5 minutes ago
SARS-CoV-2-HIV: Researchers from University of Chicago-USA and Northwestern University, Chicago-USA have in a new study uncovered the similarities and differences between SARS-CoV-2 And HIV-1 infections in terms of cellular and immune responses.
The key findings of the study show that:
-Both COVID-19 and HIV-1+ patients show disease-specific inflammatory immune signatures
-COVID-19 patients show more productive humoral responses than HIV-1+ patients
-SARS-CoV-2 elicits more enriched IFN-I signaling relative to HIV-I
-Divergent, impaired metabolic programs distinguish SARS-CoV-2 and HIV-1 infections
-COVID-19-specific IFN-I correlated genes were enriched in MAPK signaling, and p38 MAPK inhibition was previously reported to reduce human coronavirus HCoV-229E viral replication in human lung epithelial cells. Hence, inhibitors targeting MAPK signaling also have the potential to be used for treating COVID-19 patients.
Both SARS-CoV-2 and HIV-1 are RNA viruses that have killed millions of people worldwide.
It should be noted that understanding the similarities and differences between these two infections is critical for understanding disease progression and for developing effective vaccines and therapies, particularly for 38 million HIV-1+ individuals who are vulnerable to SARS-CoV-2 co-infection.
The
SARS-CoV-2-HIV study team utilized single-cell transcriptomics to perform a systematic comparison of 94,442 PBMCs from 7 COVID-19 and 9 HIV-1+ patients in an integrated immune atlas, in which 27 different cell types were identified using an accurate consensus single-cell annotation method.
Although immune cells in both cohorts show shared inflammation and disrupted mitochondrial function, COVID-19 patients exhibit stronger humoral immunity, broader IFN-I signaling, elevated Rho GTPase and mTOR pathway activities, and downregulated mitophagy.
The study findings elucidate transcriptional signatures associated with COVID-19 and HIV-1 that may reveal insights into fundamental disease biology and potential therapeutic targets to treat these viral infections.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2022.01.10.475725v1
The ongoing (COVID-19 crisis, caused by SARS-CoV-2 pathogen, has infected more than 324 million individuals and claimed more than 5.53 million lives worldwide.
At the same time, according to a current report of the Joint United Nations Programme on HIV/AIDS (UNAIDS), more than 38 million people are living with HIV-1 (PLWH) and over 36 million AIDS-related deaths have occurred since the beginning of the AIDS epidemic.
Research have shown that both SARS-CoV-2 and HIV-1 are RNA viruses and have higher mutation rates than DNA viruses. Although both the viruses have been chara
cterized as highly virulent, the manner of disease progression differs considerably. For example, in the case of SARS-CoV-2, the mortality and morbidity can be observed within a few days of infection, whereas for HIV-1 infection, it takes months or years. Also, in SARS-CoV-2 infection, neutralizing antibodies are generated rapidly post-infection, but with PLWH, antibodies take many years to develop.
To date, studies have shown that PLWH with compromised immune systems makes them susceptible to SARS-CoV-2 infection. Additionally, this group has exhibited suboptimal responses to SARS-CoV-2 vaccination. Hence, complete immune profiling of SARS-CoV-2 and HIV-1 infections would elucidate the mechanism of disease progression, which could guide researchers in the discovery of novel therapeutics.
It has also been found that many COVID-19 patients with severe infection produce high levels of inflammatory cytokines and chemokines, such as IL-6, IL-10, TNF-α, IFN-γ, and IP-10. Similarly, these cytokines are also released during acute HIV-1 infection and can persist if left untreated. Different types of immune cells drive inflammatory responses during viral infection.
Importantly the distribution and cell type-specific functions of different immune cells, such as T cells, B cells, macrophages, natural killer cells, dendritic cells, monocytes, differ across various infections, stages of disease progressions, and conditions.
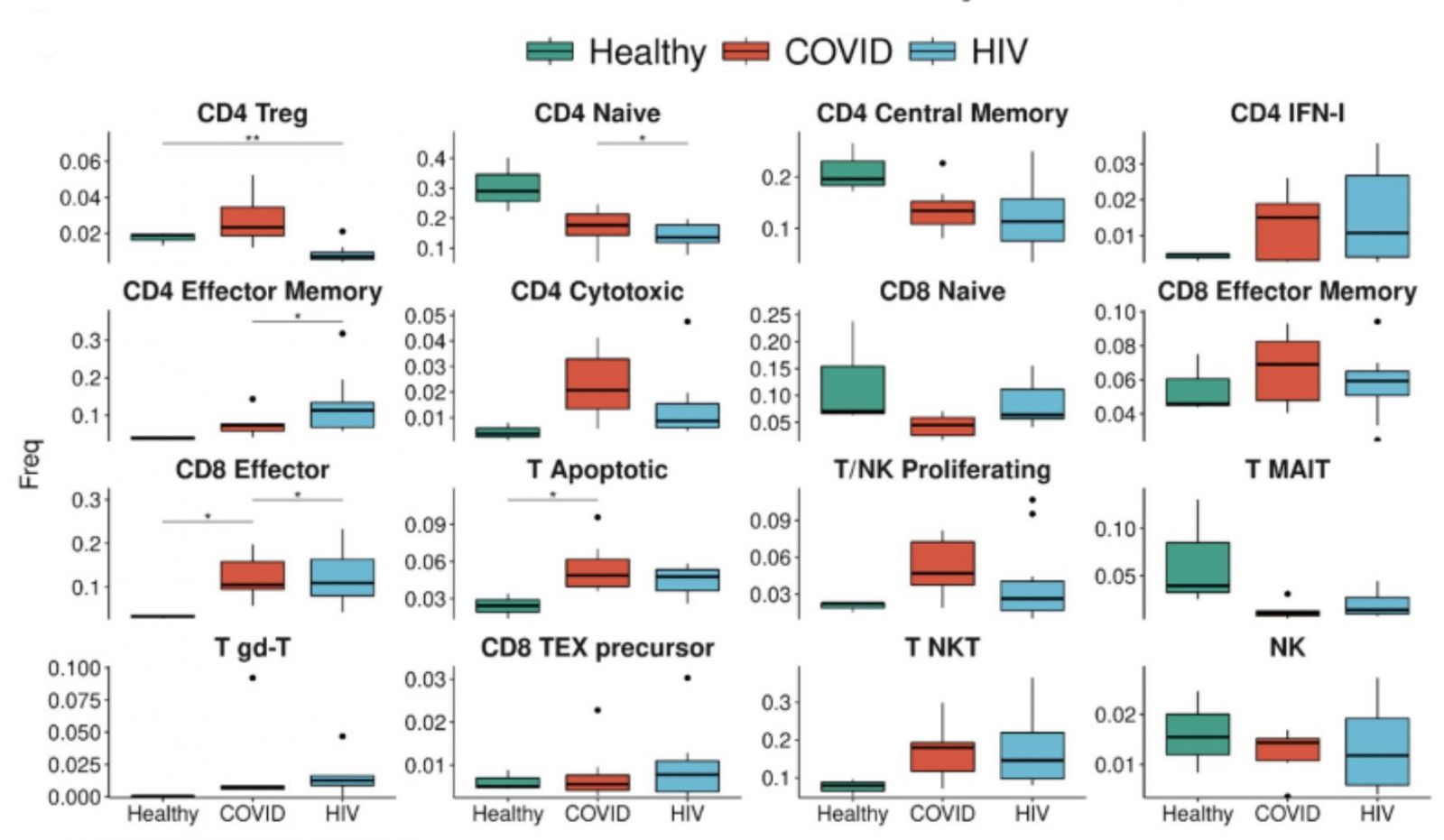 A strong signature of T cell apoptosis in COVID-19, more than HIV, and a loss of naive T cells. COVID-19 patients have an even higher proportion of IFN1 + CD4+ T cells compared to HIV-1 + patients. The enrichment of apoptotic T-cell subpopulations in both groups is consistent with the notion that HIV-1 infection leads to low levels of CD4+ T cells through pyroptosis of abortively infected T cells and apoptosis of uninfected bystander cells as well as the observation that severe COVID-19 patients frequently experienced lymphopenia.
A strong signature of T cell apoptosis in COVID-19, more than HIV, and a loss of naive T cells. COVID-19 patients have an even higher proportion of IFN1 + CD4+ T cells compared to HIV-1 + patients. The enrichment of apoptotic T-cell subpopulations in both groups is consistent with the notion that HIV-1 infection leads to low levels of CD4+ T cells through pyroptosis of abortively infected T cells and apoptosis of uninfected bystander cells as well as the observation that severe COVID-19 patients frequently experienced lymphopenia.
The process of single-cell RNA sequencing (scRNA-seq) has been extensively used to understand the heterogeneity within immune cell subsets. This sequencing method provides highly accurate annotation of individual cells. It has become a powerful tool for elucidating complex cell-cell interactions and understanding the subpopulation dynamics with single-cell resolution.
To date, may researchers have indicated that not much evidence is available regarding immune cell populations during HIV-1 and COVID-19 infections at the single-cell level.
Though several scRNA-seq atlases have been developed on COVID-19, they substantially differ in terms of granularity and markers used for annotation. Not many HIV-1 scRNA-seq profiling studies are available that are comparable to COVID-19 infection.
The study team from University of Chicago and Northwestern University had developed a single-cell atlas that presents the shared and distinct immune responses and metabolism following SARS-CoV-2 and HIV-1 infections.
The study team utilized single-cell transcriptomics to systematically compare scRNA-seq data of 115,272 single Peripheral Blood Mononuclear Cells (PBMCs) obtained from seven COVID-19, nine HIV-1, and three healthy patients.
The team managed to produce a high-quality unified cellular atlas of the immune landscape by combining the advantages of all three methods to annotate scRNA-seq data that include molecular-profile-correlation-based label transfer, manual annotation, and deep-learning-based classification.
This newly developed atlas enabled the study team to compare the phenotypic features and regulatory pathways of principal immune cells.
The study findings showed common signatures of inflammation, i.e., IFN-I and cytokine-mediated signaling, as well as errored mitochondrial function in both COVID-19 and HIV-1.
The difference between COVID-19 and HIV-1 however, was found in terms of antibody diversity, cell signaling, IFN-I signaling, and metabolic function.
For example, cytokine response by IL-2, IL-4, and IL-20 signaling was found to be more prominent in COVID-19 patients compared to HIV-1 patients that displayed high levels of NF-kB signaling.
The study team identified 27 different cell types that include five B cell subsets, two dendritic cell subsets, four monocyte subsets, seven CD4+ T cell subsets, eight CD8+ T cell subsets, and one natural killer cell subset, post COVID-19 and HIV-1 infections.
However, the types and frequencies of cellular communications among immune cells differed significantly between COVID-19 and HIV-1 patients.
The study findings reported the development of inhibitory interactions mediated by CTLA4 and HAVCR2 that were distinctive to COVID-19 patients.
In line with previous reports, a robust humoral immune response was found in both COVID-19 and HIV-1 patients. Additionally, IFN-I signaling has been reported to be closely linked with both HIV-1 and COVID-19 infection, which promotes essential cellular functions such as cell signaling, motility, and cytokine secretion.
The study team reported a decrease in mitochondrial oxidative phosphorylation (OXPHOS) and ribosome biogenesis in response to both SARS-CoV-2 and HIV-1 infection.
The study findings serve as a vital resource to understand the pathophysiological differences between COVID-19 and HIV-1. It revealed that the HIV-1 antibody repertoire was much less diverse compared to the COVID-19 antibody repertoire.
A key advantage of repertoire mapping is that it helps scientists locate high-frequency and overlapping combinations in sequences, which could inspire antibody-based therapeutics to treat comorbid patients. The study team are optimistic that this study will help develop novel molecular targets for treating these diseases.
For more about
SARS-CoV-2-HIV, keep on logging to Thailand Medical News.

 A strong signature of T cell apoptosis in COVID-19, more than HIV, and a loss of naive T cells. COVID-19 patients have an even higher proportion of IFN1 + CD4+ T cells compared to HIV-1 + patients. The enrichment of apoptotic T-cell subpopulations in both groups is consistent with the notion that HIV-1 infection leads to low levels of CD4+ T cells through pyroptosis of abortively infected T cells and apoptosis of uninfected bystander cells as well as the observation that severe COVID-19 patients frequently experienced lymphopenia.
A strong signature of T cell apoptosis in COVID-19, more than HIV, and a loss of naive T cells. COVID-19 patients have an even higher proportion of IFN1 + CD4+ T cells compared to HIV-1 + patients. The enrichment of apoptotic T-cell subpopulations in both groups is consistent with the notion that HIV-1 infection leads to low levels of CD4+ T cells through pyroptosis of abortively infected T cells and apoptosis of uninfected bystander cells as well as the observation that severe COVID-19 patients frequently experienced lymphopenia.