RXRG And SMARCA5 Identified As Regulons Modulating The Transcriptional Response To SARS-CoV-2 Infection
Nikhil Prasad Fact checked by:Thailand Medical News Team Feb 29, 2024 1 year, 1 month, 2 weeks, 4 days, 3 hours, 4 minutes ago
COVID-19 News: The relentless spread of coronavirus disease 2019 (COVID-19), fueled by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has propelled researchers into a race against time to understand the intricate molecular mechanisms governing the infection's diverse manifestations. Amidst the myriad cellular processes implicated in COVID-19 pathophysiology, including interferon (IFN) responses and the roles of ACE2 and TMPRSS2 proteins, there is a critical need to delineate the transcriptional programs orchestrating these responses. This
COVID-19 News report delves into the identification of regulons, which are clusters of co-regulated genes, as pivotal players in modulating the transcriptional response to SARS-CoV-2 infection across diverse tissues and cell types.
 Regulons Modulating The Transcriptional Response To SARS-CoV-2 Infection Identified.
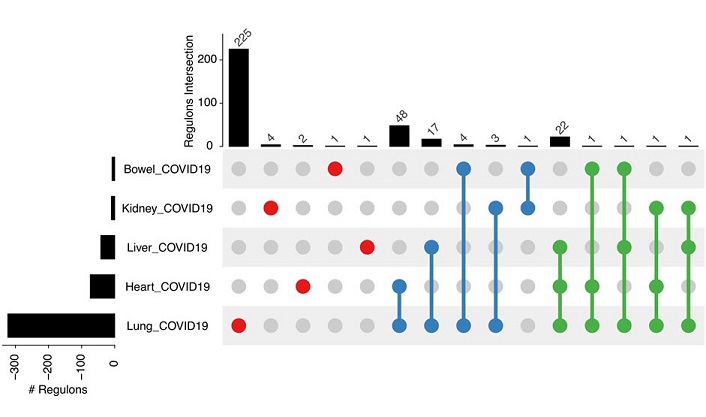
Analysis of transcriptome data for tissues with COVID-19 and controls. Upset plot of differentially active and specific regulons per experiment compared to controls.
Understanding the Basics of Regulons
Regulons Modulating The Transcriptional Response To SARS-CoV-2 Infection Identified.
Analysis of transcriptome data for tissues with COVID-19 and controls. Upset plot of differentially active and specific regulons per experiment compared to controls.
Understanding the Basics of Regulons
In the realm of molecular genetics, regulons emerge as units of genes cohesively regulated by the same regulatory gene, which expresses a protein with repressive or activating functions. These co-regulated operons, housing single or multiple contiguous genes along the genome, are governed by shared promoters and controlled by one or a set of transcriptional factors (TFs). This exploration into the world of regulons aims to shed light on the regulatory mechanisms dictating the transcriptional response to SARS-CoV-2 infection.
Collaborative Research Efforts
A collaborative effort spanning institutions such as the Universidad Nacional Autónoma de México, Fred Hutchinson Cancer Center, Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Lieber Institute for Brain Development, Johns Hopkins Bloomberg School of Public Health, Institute of Biomedical Research of Salamanca, and the Cancer Research Center has undertaken the task of unraveling the complexities of the transcriptional response to SARS-CoV-2 infection. Leveraging diverse publicly available transcriptional data, including samples from COVID-19 patients' lung autopsies and primary cells, the research team embarks on reverse engineering gene regulatory networks to delineate the regulatory mechanisms underpinning SARS-CoV-2 infection across varied tissues and cell lines.
COVID-19 Pathophysiology and Transcriptional Response
The intricate interplay between SARS-CoV-2 and human cells begins with the interaction between the viral S protein and the ACE2 receptor, facilitated by TMPRSS2. While these proteins are highly expressed in epithelial cells, their presence extends to other cell types, including myocardial, renal, enterocytes, and endothelial cells. Unraveling the molecular
intricacies of this interaction is crucial, as symptoms such as respiratory limitations and diarrhea are likely linked to the infection of epithelial cells in the lung and gut.
Within infected cells, the virus exploits the cellular machinery for replication, ultimately leading to cell death. Recognition of viral nucleic acids by pattern recognition receptors (PRRs) triggers a transcriptional response mediated by IFN regulatory factors (IRFs) and nuclear factor κB (NF-κB). While some mechanisms are shared across viral infections, the unique response triggered by SARS-CoV-2 necessitates a meticulous investigation into the molecular mechanisms at play.
The Power of Transcriptional Regulatory Networks
Transcriptional regulatory mechanisms emerge as a potent tool to decipher the cascades of information governing gene expression during SARS-CoV-2 infection. Computational approaches, rooted in gene expression data, facilitate the identification of co-regulated genes and the transcription factors orchestrating this regulation. Previous studies utilizing single-cell RNA-seq data have uncovered regulatory networks in lung epithelium-derived cell lines, offering insights into the regulating presence of TFs and the absence of tropism factors specific to SARS-CoV-2. However, a comprehensive analysis of global infection-induced regulatory networks is yet to be undertaken.
Unraveling Gene Regulatory Networks in SARS-CoV-2 Infection
This research endeavors to fill the gaps in our understanding of the gene regulatory networks deployed during the response to SARS-CoV-2 infection in humans. By analyzing published SARS-CoV-2 transcriptome infection data in cell lines and tissues, the goal is to identify regulatory interactions influencing gene expression. This approach promises to augment our current knowledge of human-SARS-CoV-2 interactions and provide crucial insights into relevant regulatory mechanisms.
Results Unveiling Unique Transcription Factors
The analysis of SARS-CoV-2 infection response in cell culture reveals the influence of specific transcription factors not found in other viral infections. Principal component analysis (PCA) of transcriptome data from various respiratory viruses demonstrates distinct clustering, with SARS-CoV-2 infections showing a unique pattern.
The identification of regulons such as RXRG and SMARCA5, unique to SARS-CoV-2 infection, suggests their potential roles in viral response, apoptosis, and NF-κB signaling. Notably, FOXP4 and ZNF730 regulons, specific to SARS-CoV-2 infection in A549 cells, hint at their relevance in understanding long COVID and warrant further exploration.
Extensive Regulatory Response in SARS-CoV-2 Lung Infection
Moving beyond cell culture, the analysis of SARS-CoV-2 lung infection regulons delves into the tissue context. Data from COVID-infected lung tissues exhibit remarkable heterogeneity, highlighting the intricate regulatory response within this vital organ. The lung emerges with the highest number of tissue-specific differentially activated (DA) regulons, indicative of an extensive regulatory response driven by efficient viral replication. Comparisons with other tissues underscore tissue-specific variations in the regulatory landscape, shedding light on potential therapeutic targets.
Discussion: Implications and Future Directions
This study significantly advances our understanding of SARS-CoV-2 infection by identifying DA regulons pivotal in orchestrating specific infection responses. The uniqueness of regulons such as RXRG, SMARCA5, FOXP4, and ZNF730 in SARS-CoV-2 infection opens avenues for further research, potentially unraveling key players in viral response, apoptosis, and long COVID. The presence of previously identified regulons related to inflammatory responses, such as IRF1 and SMARCC1, underscores the complexity of the host-virus interplay.
While the current study provides valuable insights, it is not without limitations. Future investigations incorporating single-cell data are essential to offer a more granular understanding of the regulatory landscape. Additionally, exploratory in nature, these results lay the groundwork for further experimental validations to confirm the biological relevance of identified regulons. Leveraging community-driven data and computational approaches, this study represents a crucial step towards unraveling the intricate gene regulatory networks underpinning SARS-CoV-2 infection, paving the way for targeted therapeutic interventions and a deeper comprehension of COVID-19 pathophysiology.
The study findings were published in the peer reviewed journal: Frontiers in RNA Research.
https://www.frontiersin.org/articles/10.3389/frnar.2024.1334873/full
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
