SARS-CoV-2 Exploits A Novel Mechanism To Induce Apoptosis In Respiratory Epithelial Cells Via SUD2 And Nsp5 Interaction With BclII G-Quadruplex
Latest SARS-CoV-2 Research - SUD2 - Nsp5 - BclII G-Quadruplex Mar 09, 2023 2 years, 1 month, 1 week, 4 days, 3 hours, 24 minutes ago
Latest SARS-CoV-2 Research: The COVID-19 pandemic has caused significant disruptions to society, highlighting the urgent need for effective treatments for the disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). While much research has focused on the spike and RNA-dependent RNA polymerase proteins of SARS-CoV-2, which are essential for virus entry and replication, the molecular mechanisms underlying the severe pathogenicity of the virus remain unclear.
One protein domain of interest is SUD2, which is found in the Nsp3 protein of SARS-CoV-2 and shares significant similarity with the SUD1 domain of SARS-CoV.
SUD1 has been shown to interact with G-quadruplex (G4) DNA structures, which may be responsible for its pathogenicity.
The study team from Sichuan University, Chengdu, China aimed to investigate how SUD2 interacts with G4 structures and other proteins of SARS-CoV-2 to exert its pathogenic function.
Their study findings discovered that Nsp5, also known as the main protease or 3-chymotrypsin-like protease, is a binding partner of the SUD2-N+M domains (SUD2core) with high affinity.
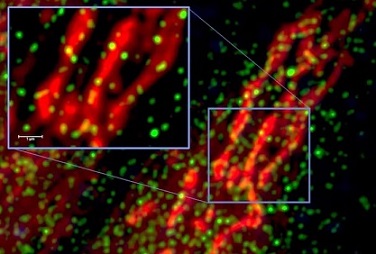
Through biochemical and fluorescent assays, the researchers demonstrated that this complex forms in the nucleus of living host cells. Additionally, the SUD2core-Nsp5 complex displayed significantly enhanced selective binding affinity for the G4 structure in the BclII promoter compared to SUD2core alone.
In order to further investigate the structural features of this complex, the researchers used AlphaFold2 and molecular dynamics analysis to predict the atomic interactions between SUD2core, Nsp5, and BclII G4 DNA.
The study team found that a positively charged groove at the top of the M domain of SUD2 interacts with a negatively charged surface of BclII G4, forming hydrogen bonds between K578, S582, and R586 of SUD2 and the phosphodiester backbone of BclII G4. This interaction downregulated the expression of the antiapoptotic gene BclII and subsequently led to augmented apoptosis of respiratory cells.
The findings of this study provide new information about the interactions between SUD2, Nsp5, and G4 DNA structures and their roles in the pathogenicity of SARS-CoV-2.
Importantly, this
Latest SARS-CoV-2 Research highlights the importance of protein-protein interactions (PPI) in the function of viral proteins and the development of effective therapies. In addition to targeting SUD2 or Nsp5, destabilizing G4 DNA structures or their mutual interfaces may also be a promising avenue for future therapies.
Past studies have shown that the induction of apoptosis in infected respiratory cells is a hallmark of severe COVID-19 disease. Therefore, understanding the molecular mechanisms underlying this process is crucial for developing effective treatments. The interaction between SUD2 and Nsp5 and their enhanced binding affinity for BclII G4 DNA may provide a possible explanation for how SARS-CoV-2 induces apoptosis in respiratory cells.
The SUD2-Nsp5 complex also has implications for the function of Nsp5 in the replication and transcription of SARS-CoV-2. As the main protease, Nsp5 cleaves t
he replicase polypeptides and is essential for virus replication. The interaction between Nsp5 and SUD2 may influence the function of Nsp5, and further research is needed to address this possibility.
The discovery of the SUD1 sequence in the genome of SARS-CoV during the 2003 epidemic sparked interest in understanding how the protein domain related to the virus's pathogenicity. Later, the SUD1 domain of SARS-CoV-2 Nsp3 was found to bind with oligo G nucleotides, especially G4 structural DNA, which may contribute to the severe pathogenicity of SARS-CoV-2. In the COVID-19 pandemic, the similar SUD2 sequence was identified in the genome of SARS-CoV-2, and it was found to interact with G4s structures in vitro, similar to SUD1.
In this study, the researchers sought to investigate the interactions of SUD2 with other Nsps in SARS-CoV-2, how these interactions influence the G4 binding property of SUD2, and how they affect the fate of host cells. They used a yeast two-hybrid (Y2H) system to screen SUD2core binding partners in SARS-CoV-2 Nsps, and Nsp5 was identified as the sole partner of SUD2core with high affinity.
The intracellular interaction between SUD2core and Nsp5 was confirmed and detected in the nucleus of respiratory epithelial cells.
The researchers validated that SUD2core tightly binds with Bcl2G4 DNA and Nsp5 enhances this interaction, resulting in reduced BclII expression and enhanced apoptosis rates of respiratory epithelial cells.
The structural features of this complex, including detailed residue interactions and the more stable conformation of this protein-G4 DNA tertiary complex, have been rationalized through theoretical modeling and MD analysis. The study also investigated the binding target, sequencing data, and bioinformatic studies, which revealed the prevalence of many G4 motifs at gene regulatory regions in the human genome.
Although the guanine tetrad serves as the common inner core unit of G4 quadruplexes, distinct topologies and local loop structures have been found for many different G4 sequences. This G4 structural diversity controls their interactions with other components and the resulting gene regulation effects.
Also, though selective binding of SUD2core with G4 sequences has been observed, considering the enormous number of G4 sequences existing at different loci of the human genome, the interaction of SUD2core with G4s in other gene regulatory regions cannot be excluded.
Regarding the complexity of transcription regulation, among these alterations induced by SUD2core, further investigation is needed to determine which ones are the direct consequence of SUD2core-Bcl2G4 interaction. In addition to SUD2core, Nsp3 contains multiple modular protein domains, and these portions have also been investigated separately and display different roles during viral infection in host cells.
For instance, the Mac1 domain removes ADP ribosylation posttranslational modifications, and the PLpro domain antagonizes MDA5-mediated type I interferon signaling to suppress the initial immune response.
Apart from being the partner of SUD2core, Nsp5 has also been previously identified as a main protease in the SARS-CoV-2 genome and is essential for cleaving the replicase polypeptides (pp1a and pp1ab) and the replication and transcription of SARS-CoV-2.
Further research is needed to address how the interaction between SUD2 and Nsp5 influences the function of Nsp5 in this role.
The study provides new information that the SUD2core-Nsp5-Bcl2G4 interaction leads to increased apoptosis of respiratory epithelial cells, which has been identified as a hallmark of severe COVID-19 disease. Based on these results, new therapies targeting SUD2, Nsp5, destabilizing G4, or their mutual interfaces can be envisioned
The study findings were published in the peer reviewed journal: Molecular And Cellular Biology.
https://journals.asm.org/doi/10.1128/mbio.03359-22
For the
Latest SARS-CoV-2 Research, keep on logging to Thailand Medical News.
