SARS-CoV-2 News: University Of Oxford Study Shows That Alpha Or B.1.1.7 Variant Exhibits Extended Viral Shedding Time! Study Also indicates That Every Variant Behaves Differently!
Source: SARS-CoV-2 News-Alpha Variant Jul 03, 2021 3 years, 9 months, 3 weeks, 2 days, 15 hours, 33 minutes ago
SARS-CoV-2 News: Public health authorities are in for rude shock as their current policies of 10 to 14 days quarantine periods to prevent the spread of the SARS-CoV-2 are now completely obsolete. A new study led by researchers from University Of Oxford that also involved scientists and doctors from the University of Glasgow and Brighton & Sussex University Hospitals NHS Trust shows that the more transmissible Alpha or B.1.1.7 variant not only replicates slowly when in the human host but also exhibits a much longer viral shedding period than other variants.

The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2021.06.29.450133v1
The British study team provided important new insights into the early stages of infection with the SARS-CoV-2 coronavirus-the agent that causes the COVID-19 disease.
.jpg) Study: Absolute quantitation of individual SARS-CoV-2 RNA molecules: a new paradigm for infection dynamics and variant differences
Study: Absolute quantitation of individual SARS-CoV-2 RNA molecules: a new paradigm for infection dynamics and variant differences
The research team’s quantitation of SARS-CoV-2 replication dynamics at the single-cell level revealed a previously unrecognized variability among cells in supporting SARS-CoV-2 replication.
The study team was also surprised to find that the SARS-CoV-2 variant of concern B.1.1.7 (Alpha) that first emerged in the UK replicates more slowly than the Victoria isolate ie an early variant that is related to the original strain isolated in Wuhan, China.
The study team says this research finding suggests that a novel mechanism contributes to the increased transmissibility of B.1.1.7.
It is already known that the SARS-CoV-2 genome consists of a positive-sense single-strand RNA of 30 kilobases that encodes a plethora of viral proteins.
The SARS-CoV-2 coronavirus primarily targets the respiratory tract and infection is mediated by the viral spike protein when it binds to the host cell receptor angiotensin-converting enzyme (ACE2).
Upon host cell entry, the first step in the replicative life cycle is the translation of the genomic RNA (gRNA) to form the replicase complex, enabling the synthesis of further positive gRNA copies. In addition, a series of shorter sub-genomic (sg)RNAs is also synthesized that encodes the spike, nucleocapsid and envelope structural proteins, as well as non-structural proteins. Despite their essential role in establishing productive infection, these early steps in the SARS-CoV-2 replicative life cycle are currently poorly understood.
Normally, analyses of viral replication are performed using “in-bulk” approaches such as reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and conventional RNA sequencing.Co-corresponding author Professor Dr Ilan Davis from the Department of Biochemistry, University of Oxford told
Thailand Medical
>News, “While very informative, these approaches lack spatial information and do not allow single-cell analyses. The kinetics of SARS-CoV-2 RNA replication and transcription during the early phase of infection are not well understood and lack quantitative, spatial and temporal information on the genesis of gRNA and sgRNAs."
It should be noted that since the COVID-19 outbreak began in Wuhan, China, in late December 2019, several variants of concern (VOC) have emerged containing mutations that increase transmissibility and confer immune escape.
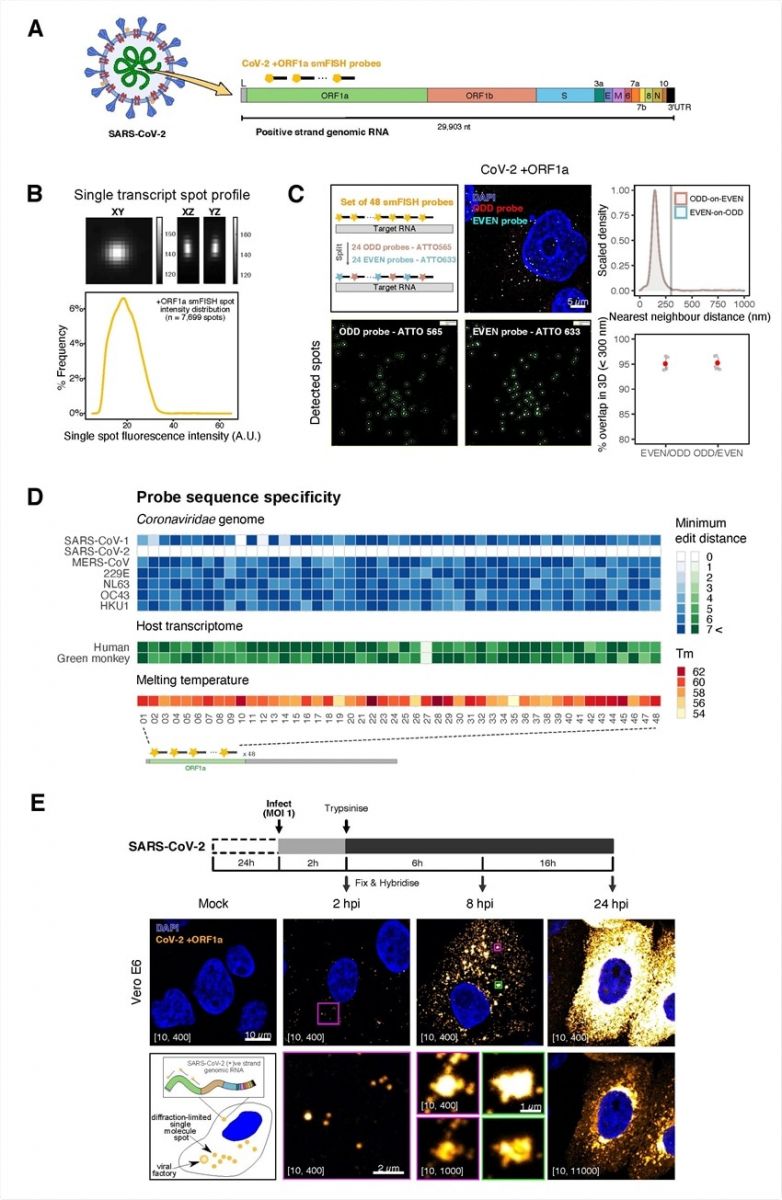 Sensitive single-molecule detection of SARS-CoV-2 genomic RNA in infected cells. (A) Schematic illustration of single-molecule fluorescence in situ hybridisation (smFISH) for detecting SARS-CoV-2 positive-strand genomic RNA (+gRNA) within infected cells. (B) Reference spatial profile of a diffraction-limited +ORF1a smFISH spot. The gradient legend represents relative fluorescence intensity (top). Frequency distribution of smFISH spot intensities, exhibiting a unimodal distribution (bottom). (C) Assessment of smFISH detection sensitivity by a dual-color co-detection method. Maximum intensity projected images and corresponding FISH-quant spot detection views of ODD and EVEN probe sets are shown. Scale bar = 5 µm. Density histogram of nearest11 neighbour distance from one spectral channel to another (top). Vertical line indicates 300 nm distance. Percentage overlap between spots detected by ODD and EVEN split probes, calculated bidirectionally (bottom). (D) Heatmap of probe sequence alignment against various Coronaviridae and host transcriptomes. Each column represents individual 20 nt +ORF1a probe sequences. The minimum edit distance represents mismatch scores, where ‘0’ indicates a perfect match. Melting temperatures of each probe at the smFISH hybridization condition are shown. (E) Experimental design for visualizing SARS-CoV-2 gRNA with smFISH at different time points after infection of Vero E6 cells. Cells were seeded on cover-glass and 24 h later, inoculated with SARS-CoV-2 (Victoria strain at MOI 1) for 2 h. Non-internalized viruses were removed by trypsin digestion and cells fixed at the timepoints shown. Representative 4 µm maximum intensity projection confocal images are shown. Numbers at the bottom left corner indicate dynamic contrast range used to display the image. Magnified view of insets in the upper panels are shown in lower panels. Scale bars = 10 µm or 2 µm.
Sensitive single-molecule detection of SARS-CoV-2 genomic RNA in infected cells. (A) Schematic illustration of single-molecule fluorescence in situ hybridisation (smFISH) for detecting SARS-CoV-2 positive-strand genomic RNA (+gRNA) within infected cells. (B) Reference spatial profile of a diffraction-limited +ORF1a smFISH spot. The gradient legend represents relative fluorescence intensity (top). Frequency distribution of smFISH spot intensities, exhibiting a unimodal distribution (bottom). (C) Assessment of smFISH detection sensitivity by a dual-color co-detection method. Maximum intensity projected images and corresponding FISH-quant spot detection views of ODD and EVEN probe sets are shown. Scale bar = 5 µm. Density histogram of nearest11 neighbour distance from one spectral channel to another (top). Vertical line indicates 300 nm distance. Percentage overlap between spots detected by ODD and EVEN split probes, calculated bidirectionally (bottom). (D) Heatmap of probe sequence alignment against various Coronaviridae and host transcriptomes. Each column represents individual 20 nt +ORF1a probe sequences. The minimum edit distance represents mismatch scores, where ‘0’ indicates a perfect match. Melting temperatures of each probe at the smFISH hybridization condition are shown. (E) Experimental design for visualizing SARS-CoV-2 gRNA with smFISH at different time points after infection of Vero E6 cells. Cells were seeded on cover-glass and 24 h later, inoculated with SARS-CoV-2 (Victoria strain at MOI 1) for 2 h. Non-internalized viruses were removed by trypsin digestion and cells fixed at the timepoints shown. Representative 4 µm maximum intensity projection confocal images are shown. Numbers at the bottom left corner indicate dynamic contrast range used to display the image. Magnified view of insets in the upper panels are shown in lower panels. Scale bars = 10 µm or 2 µm.
Dr Davis added, “Given the current status of the pandemic, there has been a global effort to understand the biology of emergent VOC with high transmission rates and possible resistance to neutralizing antibodies.”
To date the majority of studies have so far focused on mutations within the spike protein, since they can alter host cell entry and susceptibility to infection- or vaccine-induced immunity.
Dr Davis cautioned, “However, some of the mutations map to non-structural proteins, and it is thus plausible that they impact viral replication dynamics.”
The study team developed a single-molecule fluorescence in situ hybridization (smFISH) method that enabled them to visualize SARS-CoV-2 RNAs with high sensitivity and spatial precision.
Utilizing this innovative powerful approach, the study team quantified positive-sense RNA genomes with 95% detection efficiency while simultaneously visualizing negative-sense genomes, sub-genomic RNAs, and viral proteins.
Alarmingly the study team found that SARS-CoV-2 gRNA persisted in the presence of the antiviral drug remdesivir, indicating a long half-life.
The study team says this could reflect the high secondary structure of the RNA genome that may be refractory to degradation by cellular nucleases.
The research findings revealed that SARS-CoV-2 replication was highly variable between cells, with only a small proportion displaying a high burden of viral RNA.
The study team says that recent sequencing studies of SARS-CoV-2-infected bronchial cultures identified ciliated cells as the primary target. However, only a minority of these cells contained viral RNA that may reflect variation in susceptibility.
The team says that since the human respiratory tract encompasses the nasal passage, large and small airways, and bronchioles, knowledge of the specific cell types and their SARS-CoV-2 RNA burden is limited.
Dr Davis further added, “Applying smFISH to clinical biopsies and experimentally-infected animal samples will allow us to address this important question.”
Surprisingly the detailed analysis revealed that the B.1.1.7 (Alpha) variant exhibited significantly slower replication kinetics than the Victoria isolate, yielding a lower number of gRNA and sgRNA copies per cell, fewer viral replication factories, and a lower frequency of “super-permissive” cells.
The study team says a recent longitudinal study of nasopharyngeal swabs showed that B.1.1.7 was associated with longer infection times while showing similar peak viral loads to non-B.1.1.7 variants.
The researchers suggested that this extended duration of viral shedding may contribute to increased transmissibility, which would be consistent with the study data showing reduced replication of B.1.1.7 at the single-cell level.
Dr Davis said, “Replication fitness will be defined by the relationship of the virus with its host cell.”
Aggressive replication may trigger cellular antiviral sensors whereas reduced replication may enable the virus to replicate and persist for longer before host antiviral sensors are triggered.
The study team concluded “Such differences, and their impact on host antiviral responses, are likely to be of key importance for our understanding of the success of viral variants to spread through the population.”
Please help donate to sustain this website and support all our research initiatives. https://www.thailandmedical.news/p/sponsorship
For the latest
SARS-CoV-2 News, keep on logging to Thailand Medical News.

.jpg)
