SARS-CoV-2 Omicron Mutation Trajectory Is Faster Than What Has Been Seen In The Influenza Virus Over The Last 30 Years!
Nikhil Prasad Fact checked by:Thailand Medical News Team Feb 23, 2024 1 year, 2 months, 2 days, 23 hours, 56 minutes ago
COVID-19 News: Since its emergence in 2019, the SARS-CoV-2 virus has sparked a global pandemic, causing widespread infections and continually evolving through mutations. Scientists, particularly those at Akita Prefectural University in Japan, have been closely monitoring these mutations to understand their patterns and implications for the trajectory of the virus. Unlike the gradual accumulation of mutations seen in the influenza virus over the last 30 years, SARS-CoV-2 mutations exhibit a distinct pattern characterized by abrupt emergence and targeted changes, particularly in the spike protein and small open reading frames (ORFs). The latest variant, JN.1, has shown a trajectory of change that sets it apart from previous Omicron variants, raising concerns about the virus's evolving nature.
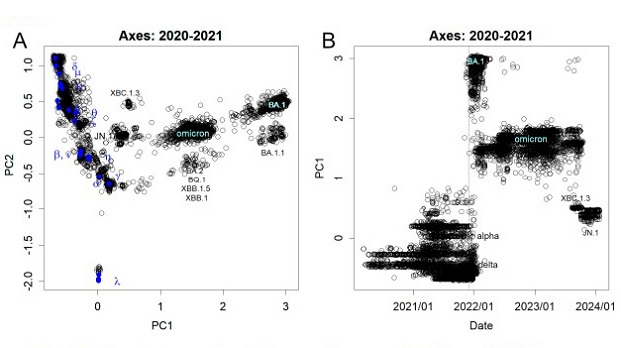 Mutations observed from nucleotide sequence. The axes were found at the end of 2021 when BA.1 emerged. (A) PC1 and PC2. BA.1 and Omicron were largely separated by mutations not seen in the previous variants, forming PC1. Some features of BA.1 were inherited by subsequent Omicron variants. However, XBC1.3 and JN.1 have reverted from the shared mutations and are closer to the former variants. (B) Time course of PC1. BA.1 appeared after 1st December 2021, indicated by the dotted line, and the former variants were almost absent. This was then replaced by a population of Omicron variants, which eventually moved to JN.1.
Understanding the Omicron Variant
Mutations observed from nucleotide sequence. The axes were found at the end of 2021 when BA.1 emerged. (A) PC1 and PC2. BA.1 and Omicron were largely separated by mutations not seen in the previous variants, forming PC1. Some features of BA.1 were inherited by subsequent Omicron variants. However, XBC1.3 and JN.1 have reverted from the shared mutations and are closer to the former variants. (B) Time course of PC1. BA.1 appeared after 1st December 2021, indicated by the dotted line, and the former variants were almost absent. This was then replaced by a population of Omicron variants, which eventually moved to JN.1.
Understanding the Omicron Variant
The Omicron variant, which surfaced in late 2021 as reported by past
COVID-19 News reports, has proven to be more transmissible than any previous variant, dominating the virus's history for about half of its existence. Researchers aimed to unravel the characteristics of the Omicron variant and compare the mutational patterns of SARS-CoV-2 with the influenza H1N1 virus. To achieve this, principal component analysis (PCA) was employed, offering a comprehensive overview and revealing distinct differences between the two viruses.
Mutational Patterns in SARS-CoV-2
The study identified a significant departure from the mutational patterns observed in influenza. Unlike influenza, where mutations accumulate gradually, SARS-CoV-2 mutations manifest abruptly and substantially. The Omicron variant, represented by BA.1, exhibited a trajectory of change distinct from previous variants, marked by sudden emergence and subsequent dominance.
The emergence of JN.1 further added to the complexity, with substantial changes at previously unmutated sites, indicating a new direction of mutation.
Sites of Mutation in the Genome
Analyzing the nucleotide sequences, researchers observed that the mutations in SARS-CoV-2 were concentrated in the spike glycoprotein (S), with frequent variations across nearly all bases. This stands in stark contrast to influenza, wher
e all ORFs mutate at uniform rates. The study also highlighted the rarity of insertions in SARS-CoV-2, with the JN.1 variant showing some unique features closer to earlier variants than other Omicrons.
Mutations Observed in Spike Protein
Further investigation into the spike protein mutations revealed a unique pattern in SARS-CoV-2, with abrupt and discrete clusters of mutations. The rapid and continuous mutations observed in the spike protein raised concerns about the practicality of targeting it for detection or immune response purposes. The study suggested that focusing on other regions, such as ORF3a, E, and specific regions of N, might offer a more effective approach, considering the rapid mutation rate of the virus.
Comparative Analysis with Influenza
The comparison between SARS-CoV-2 and influenza highlighted fundamental differences in their evolutionary patterns. Influenza undergoes continuous evolution of a single variant, accumulating mutations over time, leading to recurrent outbreaks. In contrast, SARS-CoV-2 variants, infecting fewer individuals, exhibit independent mutations, and new variants appear suddenly, irrespective of previous ones. The study emphasized the impact of the spike protein's binding to ACE2, resulting in multiple strains circulating simultaneously.
Implications for Public Health
The rapid mutation rate of SARS-CoV-2, particularly in the spike protein, has posed challenges for detection methods and immune response strategies. The study suggested that repeated vaccinations targeting the rapidly mutating spike protein may not be the most effective approach. Instead, considering the mutational patterns observed, a vaccine targeting other regions of the virus may offer longer-lasting efficacy with potentially fewer health concerns associated with repeated vaccinations.
Conclusion
The research conducted at Akita Prefectural University sheds light on the unprecedented mutational dynamics of SARS-CoV-2, particularly the Omicron variant. The distinct patterns observed in the trajectory of mutations, compared with the influenza virus, emphasize the need for a nuanced approach in understanding and addressing the challenges posed by the virus. As JN.1 and future variants continue to emerge with unique characteristics, ongoing vigilance and research are crucial for adapting public health measures and vaccine strategies to effectively combat the evolving nature of the SARS-CoV-2 virus.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.preprints.org/manuscript/202402.1283/v1
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
