SARS-CoV-2 Spike’s Fusion Peptide Binds To Negatively-Charged Phospholipids In Cell Membranes Of Host
SARS-CoV-2 Research - Fusion Peptide Mar 18, 2023 2 years, 1 month, 2 days, 8 hours, 36 minutes ago
A new study by researchers from the Institute of Research, Development, and Innovation in Healthcare Biotechnology (IDiBE), Universitas “Miguel Hernández”, Spain has found that SARS-CoV-2 spike’s fusion peptide binds to negatively-charged phospholipids in cell membranes of host without significant changes to its secondary structures.
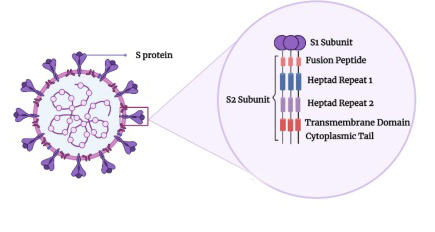
The SARS-CoV-2 virus which causes the COVID-19 disease, is a positive-sense, single-stranded RNA virus with an envelope. It surfaced in late 2019, and by early 2020, it had escalated into a global pandemic. To date, over 650 million cases and almost 7 million deaths have been reported worldwide. Despite new vaccines, the pandemic continues to significantly affect global health. One main focus in the development of antiviral treatments for COVID-19 is targeting membrane fusion, which is crucial for the virus to enter host cells.
The S2 subunit of the virus's Spike protein, a class I membrane fusion protein, houses the fusion domain directly involved in the fusion process. Understanding this mechanism at a molecular level is key for creating effective antiviral approaches.
The study team employed all-atom molecular dynamics to examine the binding of the SARS-CoV-2 fusion peptide to specific phospholipids in model membranes composed of a single phospholipid and cholesterol in the presence of either Na+ or Ca2+ ions.
The study findings revealed that the fusion peptide can bind to the membrane without significant changes to its secondary structure. It tends to favor electronegatively charged phospholipids and does not bind to cholesterol. Gaining insight into the membrane fusion process and the molecular interactions at play will aid in developing antiviral compounds to more effectively combat such viruses.
The study findings were published in the peer reviewed journal: Membranes.
https://www.mdpi.com/2077-0375/13/3/344
Typically, coronaviruses (CoVs) have main structural components such as spike (S), membrane (M), envelope (E) proteins, nucleoprotein complexed with RNA, and the membrane envelope.
The S protein, a class I membrane fusion protein, is crucial for SARS-CoV-2 infectivity by binding to the receptor and facilitating host cell entry via membrane fusion. The S protein forms a homotrimer on the viral membrane, giving CoVs their crown-like appearance. It has two subunits: S1, containing the receptor binding domain, and S2, housing the fusion domain (fusion peptide, FP). The S2 subunit, where the FP resides, is highly conserved.
Membrane fusion is initiated by conformational changes in the S protein that expose the FP, allowing it to interact with the host lipid bilayer. The exact FP sequence in coronaviruses remains unidentified, but two well-conserved sequences have been suggested: FP1 and FP2. These sequences possess conserved motifs and interact with specific membrane lipids. Both FP1 and FP2 seem necessary for fusion and bind the membrane independently, possibly acting cooperatively and promoting each other's membrane binding. The preferential binding of each sequence may depend on lipid composition and specificity, ultimately resulting in all FP sequences binding and inserting into the membrane.
The structure of the fusion peptide (FP), a hydrophobic and intrinsically disordered component, is critical for understanding membrane fusion at the mole
cular level and developing effective antiviral strategies.
Currently, there is limited information on FP's structure and dynamics in membrane insertion, interaction, and modulation leading to viral-cell membrane fusion. Membrane fusion is not a spontaneous process and requires overcoming energy barriers.
This study used molecular dynamics (MD) simulations to investigate the binding and interaction of SARS-CoV-2 FP with specific phospholipids in a model membrane. The study team found that the FP interacts more strongly with certain negatively-charged phospholipids, but not with cholesterol. These interactions could be crucial for the initial stages of membrane fusion, potentially followed by FP interaction with cholesterol at a deeper level. The secondary structure of FP remained stable during binding, regardless of the membrane system or ions present.
The binding process seems to involve a combination of charged and hydrophobic residues, optimizing FP's interactions with phospholipids. Several factors contribute to reducing the energy required for membrane fusion, including specific interactions between protein residues and phospholipid head-groups, physical changes in the membrane, and the formation of lipid-enriched domains.
The study team concluded, “The entry of SARS-CoV-2 into cells occurs through membrane fusion, a critical mechanism and a primary target for developing new antiviral therapies. The S2 subunit of the SARS-CoV-2 spike protein contains the fusion domain, which binds and interacts with lipids in the membrane. Using molecular dynamics, this study examined the interaction between the fusion domain and phospholipids in a model membrane, finding a specific interaction with negatively charged phospholipids. Both hydrophobic and charged amino acids are involved in membrane binding, suggesting that a combination of hydrophobic and electrostatic effects drive fusion, enabling close association with membrane phospholipids. This binding allows for the formation of domains in the membrane that facilitate interactions between other parts of the SARS-CoV-2 spike protein and the host membrane. In conclusion, the fusion domain's charge, hydrophobicity, binding to negatively charged phospholipids, and unique wrapping are essential for initiating membrane fusion, ultimately leading to the collapse of viral and cellular membranes.”
For the latest
SARS-CoV-2 Research, keep on logging to Thailand
Medical News.
