Nikhil Prasad Fact checked by:Thailand Medical News Team Jul 30, 2024 1 year, 5 months, 5 days, 22 hours, 7 minutes ago
COVID-19 News: Introduction to SARS-CoV Virulence Mechanisms
A recent study delves into how the severe acute respiratory syndrome coronavirus (SARS-CoV) increases its virulence through specific protein interactions. Researchers from the Centro Nacional de Biotecnología (CNB-CSIC) and the Universidad Católica de Valencia in Spain investigated how the virus's PDZ-binding motifs (PBMs) interact with cellular PDZ domains, leading to heightened virus virulence. This
COVID-19 News report explores the key findings of their study, shedding light on the mechanisms that make SARS-CoV so dangerous.
 SARS-CoV-2 virulence is enhanced by key protein interactions
The Role of PDZ-Binding Motifs
SARS-CoV-2 virulence is enhanced by key protein interactions
The Role of PDZ-Binding Motifs
Coronaviruses, particularly SARS-CoV, have become a significant public health concern due to their capacity to cause severe diseases like SARS and COVID-19. An important area of research has been the study of specific protein interactions between the virus and host cells. One such interaction involves the PDZ-binding motifs (PBMs) found in viral proteins and the PDZ domains in host cellular proteins.
PDZ domains are small protein interaction modules found in various organisms, from bacteria to humans. They play a critical role in signaling pathways, regulating cell proliferation, differentiation, and apoptosis. The SARS-CoV virus contains PBMs in its 3a and envelope (E) proteins. These PBMs can bind to over 400 cellular proteins with PDZ domains, significantly influencing cell function.
Key Findings on Virus Viability and Replication
The researchers conducted a comparative study on the functional motifs within the SARS-CoV 3a and E proteins. They discovered that both proteins are necessary for maximum virus replication and virulence. The study revealed that a virus missing both the 3a-PBM and E-PBM sequences was not viable. However, the presence of at least one of these proteins with a functional PBM restored the virus's viability.
Interestingly, the E protein PBM was found to be essential for virulence. The interaction between the E protein PBM and the PDZ domain of the cellular protein Syntenin-1 activates the p38-MAPK pathway, leading to increased virus pathogenicity. In contrast, the 3a protein PBM does not bind to Syntenin-1 and does not influence virus pathogenicity.
Significance of the E Protein's PBM
The E protein's PBM is vital for the virus's ability to cause severe disease. The study demonstrated that the PBM's interaction with the PDZ domain of Syntenin-1 leads to conformational changes in both PDZ domains of the Syntenin-1 homodimer. These changes are likely to influence downstream signaling pathways, contributing to the virus's pathogenicity.
Furthermore, the study revealed that specific amino acid residues in the E protein's PBM are critical for its function. The researchers performed alanine and glycine scanning mutagenesis, identifying residues 20 and 22 as necessary for optimal virus replication. Mut
ants with these residues altered showed significantly lower viral titers in cell cultures and mouse lungs.
Insights into Protein Interactions
The study used a combination of in vitro, in vivo, and in silico techniques to examine the effect of PBM binding to PDZ-containing proteins. The findings provide new insights into SARS-CoV pathogenicity mechanisms and may have implications for developing antiviral therapies. For instance, the E protein's absence fully attenuates the virus, highlighting its crucial role in virulence.
Further analysis revealed that specific amino acids within the E protein PBM are necessary for optimal virus replication. Mutagenesis analysis and in silico modeling identified that residues 20 and 22 from the carboxy-terminus of the E protein significantly impact virus replication. These residues are involved in interactions that lead to conformational changes in the PDZ domains of Syntenin-1, affecting the virus's pathogenicity.
Detailed Examination of Virus Mutants
To better understand the role of the PBM sequence in virus viability and virulence, the researchers created several virus mutants. These mutants either had the PBM sequences deleted or replaced in the 3a and E proteins. The study found that deleting both PBM sequences reduced viral titers by around tenfold, but the mutants remained viable. However, viruses with the E protein PBM deleted showed significantly reduced virulence, emphasizing the E protein PBM's critical role.
When the E protein PBM was replaced with the 3a protein PBM, the virus's virulence remained similar to the wild-type virus, suggesting that the sequence context of the PBM is vital for its biological activity. This indicates a hierarchy between the 3a and E proteins, where the E protein's PBM is more influential in determining virulence.
Structural Insights from In Silico Modeling
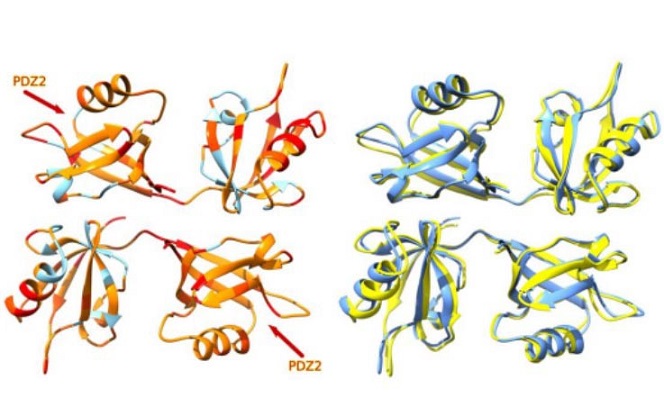
The study also explored the structural aspects of the PBM-PDZ interactions using in silico modeling. The researchers predicted that the binding of the E protein PBM to the PDZ2 domain of Syntenin-1 induces conformational changes in both PDZ domains. This binding opens the PDZ2 pocket by approximately 2 Å on average and the unbound PDZ1 pocket by about 1.3 Å, suggesting a cooperative interaction between the PDZ domains.
Potential Implications for Antiviral Therapies
These findings highlight the complexity of PBM-PDZ interactions and their impact on virus pathogenicity. The research provides valuable insights into the mechanisms by which SARS-CoV manipulates host cell functions to enhance its virulence. Understanding these interactions opens up new possibilities for developing targeted antiviral therapies that disrupt these critical protein interactions.
Conclusion
The interaction between SARS-CoV PBMs and cellular PDZ domains plays a significant role in virus virulence. The study found that the E protein PBM is essential for virulence, whereas the 3a protein PBM does not significantly affect pathogenicity. The detailed analysis of PBM interactions with cellular proteins provides a deeper understanding of SARS-CoV's mechanisms of pathogenicity, which could be crucial for developing effective antiviral strategies.
The study findings were published in the peer-reviewed journal: Viruses.
https://www.mdpi.com/1999-4915/16/8/1214
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
Read Also:
https://www.thailandmedical.news/news/viruses-use-short-sequence-motifs-to-subvert-pdz-domain-mediated-host-cell-polarity-signaling--sars-cov-2-does-this-using-the-e-proteins
https://www.thailandmedical.news/news/covid-19-news-french-study-finds-that-sars-cov-2-envelop-protein-has-a-carboxy-terminal-that-contains-a-pdz-binding-motif-that-is-pathogenetic
https://x.com/ThailandMedicaX
