South Korean Researchers Warn That SARS-CoV-2 Spike Protein Induces Lung Cancer Migration And Invasion In A TLR2-Dependent Manner!
Nikhil Prasad Fact checked by:Thailand Medical News Team Sep 14, 2023 2 years, 3 months, 5 days, 23 hours, 47 minutes ago
COVID-19 News: The ongoing COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has brought significant challenges to healthcare systems worldwide. Among the many questions surrounding this virus, its impact on individuals with pre-existing medical conditions has been a major concern. In particular, researchers at South Korea's Sungkyunkwan University School of Medicine have been investigating the relationship between SARS-CoV-2 and lung cancer. Their findings suggest that the SARS-CoV-2 Spike (S) protein may induce lung cancer migration and invasion in a Toll-like receptor 2 (TLR2)-dependent manner, shedding light on a potential link between COVID-19 and lung cancer progression.
It should be noted that many studies and
COVID-19 News reports have already shown that SARS-CoV-2 is able to either cause the progression of various cancers or is able to also cause the actual onset and development of cancers as well! (Please use the search function of Thailand Medical News which is a repository of more than 27,000 carefully selected articles and reviews of various COVID-19 studies and developments.)
Understanding the Impact of SARS-CoV-2 on Lung Cancer
Patients with lung cancer have been found to be at a higher risk of experiencing severe COVID-19 symptoms, including hospitalization and death. This heightened susceptibility has prompted researchers to investigate the molecular and cellular mechanisms underlying this phenomenon.
One key factor in this investigation is the SARS-CoV-2 Spike protein, which has been known to trigger hyper-inflammation in both epithelial cells and macrophages through Toll-like receptor (TLR) signaling pathways. Specifically, TLR1/2 and TLR2/6-dependent nuclear factor-kappaB (NF-κB) pathways have been implicated in this process.
To gain insight into the relationship between SARS-CoV-2 susceptibility and lung cancer severity, the study team analyzed microarray data from non-small cell lung cancer (NSCLC) patients. They focused on the differential expression of angiotensin-converting enzyme 2 (ACE2) and TLR2 between lung tumor tissues and matched lung normal tissues. ACE2 is associated with SARS-CoV-2 infection, while TLR2 is linked to the hyper-immune response.
The analysis revealed that lung tumor tissues with upregulated ACE2 and TLR2 exhibited significant enrichment in gene sets related to cancer progression, SARS-CoV-2 infection, and TLR signaling. This suggests a potential connection between elevated ACE2 and TLR2 expression in lung cancer tissues and disease severity.
The Role of TMPRSS2 and TLR1/6
To further investigate this connection, the study team examined the expression of other factors involved in SARS-CoV-2 infection, including transmembrane protease serine subtype 2 (TMPRSS2), TLR1, and TLR6. These proteins functionally form TLR1/2 or TLR2/6 heterodimers, contributing to the virus's virulence and pathogenesis.
The analysis identified gene sets associated with cancer progression and anti-apoptosis or inflammatory responses that were significantly enriched in lung tumor tissues with elevated TMPRSS2, TLR1, TLR2, and TLR6 expression.
Additionally, gene sets related to innate immune signaling pathways, such as TLR cascades and the nucleotide-binding and oligomerization domain (NOD)-like receptor pathway, were enriched in these
tissues. These findings indicate a potential interplay between SARS-CoV-2-related factors and lung cancer progression.
Inflammatory Cytokines and Chemokines
The SARS-CoV-2 virus has been known to induce the production of inflammatory cytokines and chemokines through TLR2-dependent activation of the NF-κB pathway. Researchers found that lung tumor tissues with elevated expression of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 were associated with gene sets related to inflammatory cytokines and chemokines, further highlighting the potential connection between SARS-CoV-2 susceptibility and lung cancer severity.
The Functional Role of the SARS-CoV-2 S Protein
To directly assess the impact of the SARS-CoV-2 Spike protein on lung cancer progression, researchers conducted experiments on lung cancer cell lines. They treated A549 and H1299 lung cancer cells with the SARS-CoV-2 S protein, as well as TLR1/2 and TLR2/6 agonists. The results were striking.
Treatment with the SARS-CoV-2 S protein, Pam3CSK4 (a TLR1/2 agonist), or FSL-1 (a TLR2/6 agonist) significantly enhanced the migration and invasion abilities of the lung cancer cells. Additionally, colony-forming and cell proliferation assays demonstrated increased growth in response to these treatments.
These findings were accompanied by increased phosphorylation of key proteins involved in the NF-κB pathway, including P65 and inhibitor of nuclear factor-κB kinases (IKKs), as well as the extracellular signal-regulated kinase (ERK). Furthermore, cytokine levels, including IL-6, IL-1β, TNF-α, and NF-κB activity, were elevated upon treatment with the SARS-CoV-2 S protein and TLR agonists.
TLR2-Dependent Activation of NF-κB
The research team sought to confirm that the observed effects were indeed TLR2-dependent. To do so, they generated TLR2-knockout (KO) lung cancer cells using the CRISPR-Cas9 gene-editing method.
In their experiments, the control lung cancer cells showed enhanced migration and invasion when exposed to the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1. However, the TLR2-KO lung cancer cells exhibited significantly reduced migration and invasion, indicating the critical role of TLR2 in these processes.
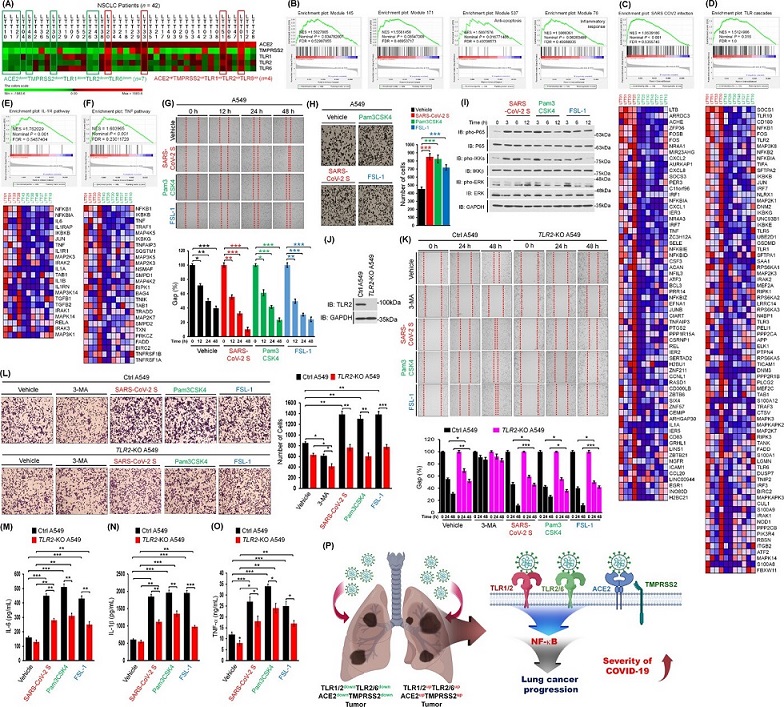 SARS-CoV-2 spike protein induces lung cancer migration and invasion in a TLR2-dependent manner. (A) 42 lung tumor tissues of NSCLC patients were listed according to the ∆Mag of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 expression in lung tumor tissues (LTTs) versus matched lung normal tissues (mLNTs). Eleven lung tumor tissues, four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues, were selected for GSEA. The color scale indicates ∆Mag value. (B) GSEA was performed in four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues indicated in (A). Gene sets for four cancer modules are presented. (C-F) Gene sets for ACE2 expressing cells with SARS-CoV-2 infection (C), TLR cascades (D), IL-1R pathway (E), and TNF pathway (F) are presented along with heat maps showing differential gene expression patterns between four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues. NES, nominal P-value, and FDR q-values are indicated in the inner panel. (G) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (H) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (I) A549 lung cancer cells were stimulated with SARS-CoV-2 S protein, Pam3CSK4, or FSL-1. Phosphorylation levels of P65, IKKs, and ERK were measured by Western blotting. (J) TLR2-knockout (TLR2-KO) A549 cells were generated using CRISPR-Cas9 gene-editing method. TLR2 expression was verified by western blotting with anti-TLR2 and anti-GAPDH (control). (K) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for different time periods (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (L) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (M-O) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 12 h. Production levels of IL-6 (M), IL-1β (N), and TNF-α (O) in culture supernatant were measured by ELISA. Results are presented as mean ± SD of three independent experiments. (P) A schematic model of how the SARS-CoV-2 virus critically affects the susceptibility to SARS-CoV-2 infection and the severity of SARS-CoV-2 infection in lung cancer patients with up-regulation of ACE2, TMPRSS2, TLR1, TLR2, and TLR6. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
SARS-CoV-2 spike protein induces lung cancer migration and invasion in a TLR2-dependent manner. (A) 42 lung tumor tissues of NSCLC patients were listed according to the ∆Mag of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 expression in lung tumor tissues (LTTs) versus matched lung normal tissues (mLNTs). Eleven lung tumor tissues, four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues, were selected for GSEA. The color scale indicates ∆Mag value. (B) GSEA was performed in four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues indicated in (A). Gene sets for four cancer modules are presented. (C-F) Gene sets for ACE2 expressing cells with SARS-CoV-2 infection (C), TLR cascades (D), IL-1R pathway (E), and TNF pathway (F) are presented along with heat maps showing differential gene expression patterns between four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues. NES, nominal P-value, and FDR q-values are indicated in the inner panel. (G) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (H) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (I) A549 lung cancer cells were stimulated with SARS-CoV-2 S protein, Pam3CSK4, or FSL-1. Phosphorylation levels of P65, IKKs, and ERK were measured by Western blotting. (J) TLR2-knockout (TLR2-KO) A549 cells were generated using CRISPR-Cas9 gene-editing method. TLR2 expression was verified by western blotting with anti-TLR2 and anti-GAPDH (control). (K) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for different time periods (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (L) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (M-O) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 12 h. Production levels of IL-6 (M), IL-1β (N), and TNF-α (O) in culture supernatant were measured by ELISA. Results are presented as mean ± SD of three independent experiments. (P) A schematic model of how the SARS-CoV-2 virus critically affects the susceptibility to SARS-CoV-2 infection and the severity of SARS-CoV-2 infection in lung cancer patients with up-regulation of ACE2, TMPRSS2, TLR1, TLR2, and TLR6. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Moreover, the use of 3-methyladenine (3-MA), an inhibitor of phosphoinositide-3-kinase (PI3K), further inhibited the migration and invasion of both control and TLR2-KO lung cancer cells.
Additionally, TLR2-KO lung cancer cells displayed reduced production of inflammatory cytokines (IL-6, IL-1β, and TNF-α) compared to control cells when exposed to the SARS-CoV-2 S protein and TLR agonists.
Conclusion
In conclusion, the research conducted at Sungkyunkwan University School of Medicine in South Korea provides valuable insights into the potential link between SARS-CoV-2 and lung cancer progression. The data suggests that individuals with upregulated ACE2, TMPRSS2, TLR1, TLR2, and TLR6 in lung cancer tissues may be more susceptible to SARS-CoV-2 infection and experience more severe disease outcomes. Furthermore, the study highlights the role of TLR2-dependent activation of NF-κB in promoting lung cancer progression in response to the SARS-CoV-2 S protein.
However, it is important to note that the role of TLR2 in lung tumor progression remains controversial, as it may also play a tumor-suppressive role in early-stage lung cancer. Further research is needed to unravel the complex molecular and cellular mechanisms behind TLR2's involvement in different stages of lung cancer. Nonetheless, this study contributes significantly to our understanding of how SARS-CoV-2 infection can influence lung cancer progression and may aid in identifying susceptibility factors in lung cancer patients.
The study findings were published as a correspondence in the peer reviewed journal: Cancer Communications.
https://onlinelibrary.wiley.com/doi/10.1002/cac2.12485
For the latest
COVID-19 News, keep on logging to Thailand Medical News
