Spanish Study Finds That SARS-CoV-2 Alters Landscape Of Host Small Non-Coding RNAs
Nikhil Prasad Fact checked by:Thailand Medical News Team Apr 18, 2024 1 year, 1 week, 4 days, 21 hours, 25 minutes ago
COVID-19 News: The ongoing COVID-19 pandemic has underscored the critical importance of understanding how viruses like SARS-CoV-2 interact with host cells at a molecular level. A recent study conducted at the University of Valencia in Spain that is covered in this
COVID-19 News report, delved into this intricate relationship, focusing specifically on how the virus remodels the landscape of small non-coding RNAs (sncRNA) within host cells. This investigation is part of a broader effort to gain insights into the molecular mechanisms underlying COVID-19 pathogenesis and to identify potential avenues for diagnostic and therapeutic interventions.
 SARS-CoV-2 Alters Landscape Of Host Small Non-Coding RNAs
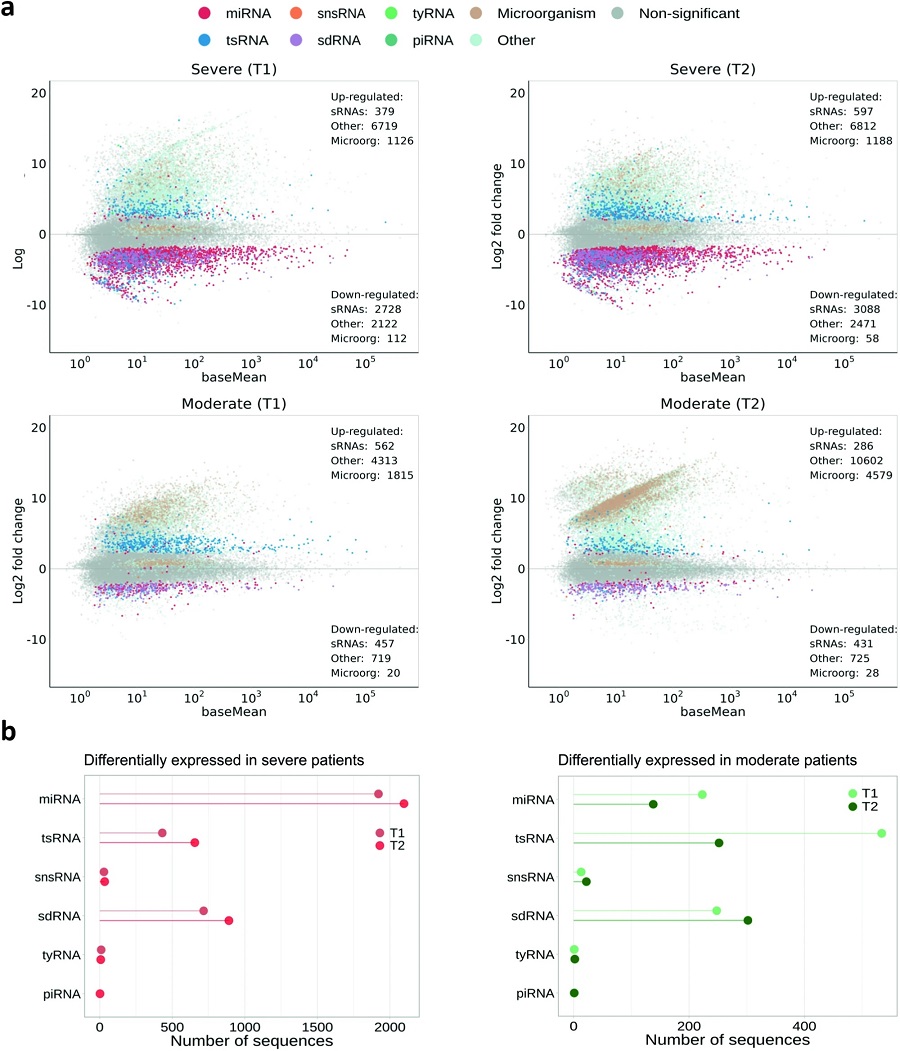
a Graphic representation of the expression values (DESeq2) of sRNA sequences for the different conditions against control samples. Each dot corresponds to a given sRNA expression value. Colors indicate significant differential expression with |log2FC | > 0.585 and FDR < 0.05 for the different sRNA families (sRNAs with non-significant differential expression are in gray). The category Other is used to label those endogenous sRNAs that could not be annotated or were annotated as miRNAs with a length not compressed between 19 and 24 nt, lncRNAs, rRNAs, scaRNAs, protein-coding or miscellaneous RNAs. sRNAs annotated as derived from microorganisms are colored in light brown. b Detail of the number of sequences differentially expressed for each sRNA family in the four conditions analyzed. miRNA micro RNA, tsRNA tRNA-derived small RNA, snsRNA small nuclear-derived RNA, sdRNA small nucleolar-derived RNAs, tyRNA tiny RNA and piRNA: Piwi-interacting RNA.
The Landscape of Small Non-Coding RNAs
SARS-CoV-2 Alters Landscape Of Host Small Non-Coding RNAs
a Graphic representation of the expression values (DESeq2) of sRNA sequences for the different conditions against control samples. Each dot corresponds to a given sRNA expression value. Colors indicate significant differential expression with |log2FC | > 0.585 and FDR < 0.05 for the different sRNA families (sRNAs with non-significant differential expression are in gray). The category Other is used to label those endogenous sRNAs that could not be annotated or were annotated as miRNAs with a length not compressed between 19 and 24 nt, lncRNAs, rRNAs, scaRNAs, protein-coding or miscellaneous RNAs. sRNAs annotated as derived from microorganisms are colored in light brown. b Detail of the number of sequences differentially expressed for each sRNA family in the four conditions analyzed. miRNA micro RNA, tsRNA tRNA-derived small RNA, snsRNA small nuclear-derived RNA, sdRNA small nucleolar-derived RNAs, tyRNA tiny RNA and piRNA: Piwi-interacting RNA.
The Landscape of Small Non-Coding RNAs
Small non-coding RNAs (sncRNAs) play crucial roles in gene regulation across various organisms. These include microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and several emerging categories such as transfer RNA-derived small RNAs (tsRNAs), small nucleolar RNA-derived small RNAs (sdRNAs), and trimmed forms of miRNAs known as tiny RNAs (tyRNAs). Despite initial perceptions of sncRNAs as byproducts of RNA metabolism, their functional significance is now widely recognized, making their study increasingly important.
While initially thought to be byproducts of RNA metabolism, sncRNAs are now recognized for their regulatory functions across various biological processes.
MiRNAs, generated from structured RNA precursors via ribonuclease enzymes, regulate gene expression through RNA silencing mechanisms. PiRNAs are involved in transposon silencing and genome stability. TsRNAs, derived from transfer RNAs, have roles in stress responses and regulation of diverse cellular processes. SdRNAs act as guides for protein interactions, while tyRNAs are implicated in alternative RNA cleavage pathways. Understanding thei
r biogenesis and functions is crucial for deciphering their roles in viral infections like COVID-19.
Impact of SARS-CoV-2 on Host sncRNAs
The study conducted at the University of Valencia utilized high-throughput RNA sequencing to analyze nasopharyngeal samples from COVID-19 patients at different stages of infection. This comprehensive approach revealed a global alteration of the sncRNA landscape in response to SARS-CoV-2 infection.
Notably, miRNA expression was predominantly down-regulated, particularly in patients with severe symptoms, highlighting a potential link between miRNA dysregulation and disease severity.
Among the most altered sncRNAs were tsRNAs derived from specific transfer RNA precursors, particularly Glu- and Gly-tRNAs. These tsRNAs, especially 5’ tRNA halves, showed significant accumulation in response to infection, suggesting their potential as biomarkers for viral presence and disease prognosis. Additionally, the study identified a down-regulation of C/D-box sdRNAs, further emphasizing the broad impact of SARS-CoV-2 on host sncRNA populations.
Clinical Implications and Future Directions
Understanding how SARS-CoV-2 alters the host sncRNA landscape carries significant clinical implications. Differential expression patterns of miRNAs, tsRNAs, sdRNAs, and other sncRNAs could serve as diagnostic markers for COVID-19 severity and progression. Moreover, elucidating the regulatory roles of these altered sncRNAs may unveil novel therapeutic targets for combating viral infections and mitigating disease outcomes.
The study's findings contribute to a growing body of research aimed at deciphering the intricate interplay between viruses and host cellular machinery. By integrating advanced sequencing techniques with bioinformatics analyses, researchers can uncover hidden regulatory networks and molecular pathways involved in viral pathogenesis. This knowledge not only enhances our understanding of COVID-19 but also paves the way for the development of precision medicine approaches tailored to individual patient profiles.
Moving forward, further investigations are warranted to validate the functional roles of specific sncRNAs identified in this study. Experimental validation using cell culture models and animal studies can provide mechanistic insights into how these sncRNAs modulate viral infection and host immune responses. Additionally, longitudinal studies involving larger patient cohorts across diverse populations can refine our understanding of sncRNA dynamics during the course of COVID-19 and its implications for long-term health outcomes.
Conclusion
In conclusion, the study conducted at the University of Valencia sheds light on the complex alterations in host sncRNAs induced by SARS-CoV-2 infection. By unraveling the regulatory landscape of miRNAs, tsRNAs, sdRNAs, and other sncRNAs, researchers gain valuable insights into the molecular mechanisms underpinning COVID-19 pathogenesis. This knowledge not only expands our fundamental understanding of virus-host interactions but also opens avenues for developing novel diagnostic tools and targeted therapies for combating COVID-19 and related viral infections.
The study findings were published in the peer reviewed journal: NPJ Systems Biology and Applications (Nature).
https://www.nature.com/articles/s41540-024-00367-z
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
