Spanish Study Reveals Genetic Variants Linked To SARS-CoV-2 Viremia And COVID-19 Pathogenesis
Nikhil Prasad Fact checked by:Thailand Medical News Team Sep 24, 2023 1 year, 6 months, 3 weeks, 3 days, 9 hours, 24 minutes ago
COVID-19 News: As the world grapples with the ongoing COVID-19 pandemic, scientific research continues to uncover new insights into the virus and its impact on human health. Nearly three years after the outbreak of SARS-CoV-2, the virus responsible for COVID-19, the global community has witnessed over 780 million infections and extensive vaccination efforts. Despite these efforts, SARS-CoV-2 continues to circulate, resulting in around 1,500 deaths per week worldwide. The diverse clinical manifestations of COVID-19 have driven scientists to explore various biomarkers that could help identify individuals at a higher risk of severe respiratory complications and death. While numerous clinical factors and biomarkers such as age, hypertension, Interleukin-6 (IL-6), and D-dimer levels have been associated with COVID-19 severity, accurately predicting disease progression and identifying predisposing conditions remain significant challenges for healthcare professionals.
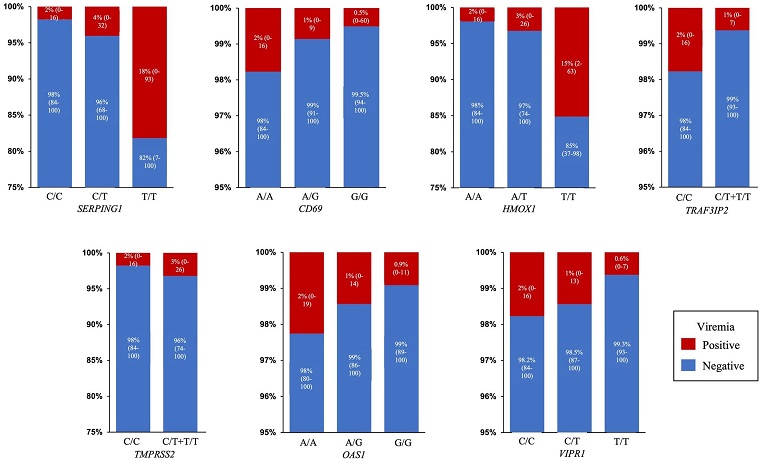 Predicted probability of viremia. Percentage and 95% Confidence Interval of predicted probability of viremia for each SNP genotype in the final model: SERPING1 (rs78958998), CD69 (rs11052877), HMOX1 (rs2071746), TRAF3IP2 (rs33980500), TMPRSS2 (rs713400), OAS1 (rs2660) and VIPR1 (rs896).
Predicted probability of viremia. Percentage and 95% Confidence Interval of predicted probability of viremia for each SNP genotype in the final model: SERPING1 (rs78958998), CD69 (rs11052877), HMOX1 (rs2071746), TRAF3IP2 (rs33980500), TMPRSS2 (rs713400), OAS1 (rs2660) and VIPR1 (rs896).
Among these potential biomarkers, the presence of SARS-CoV-2 RNA in peripheral blood, known as viremia, has emerged as a significant factor linked to severe COVID-19 outcomes. Previous studies conducted by our research group have demonstrated that patients with detectable SARS-CoV-2 viremia face a higher risk of mortality or admission to intensive care units (ICUs). Furthermore, meta-analyses and other studies have supported these findings by establishing a correlation between SARS-CoV-2 viremia and worse COVID-19 prognoses. Viremia is associated with heightened inflammatory responses, including elevated levels of C-reactive protein and IL-6. A proteomic study conducted by Li et al. also revealed that patients with viremia exhibited increased expression of SARS-CoV-2 entry factors like ACE2, CTSL, and FURIN, proinflammatory markers such as IL-6, as well as markers indicative of tissue damage and coagulation. However, the precise mechanisms and underlying genetic factors contributing to viremia remain incompletely understood.
Numerous studies have examined the association between genetic variants and COVID-19 outcomes through Single Nucleotide Polymorphism (SNP) genotyping and Genome-Wide Association Studies (GWAS) and reported these findings in various past
COVID-19 News coverages. Genes like ACE2 and TMPRSS2, which play roles in SARS-CoV-2 entry into host cells, have been extensively studied, with some of their genetic variants associated with COVID-19 severity and infectivity.
Additionally, other genomic regions related to COVID-19 severity have been identified, including those linked to the ABO blood group system and the antiviral response (OAS1, OAS2, OAS3, TYK2, IFNAR2, and IL-10). These studies have highlighted the importance of genetic factors in shaping the course of COVID-19. However, despite the potential role of genetic variants in influencing viral entry and dissemination, no prior investigation has explored the relationship between genetic variants in genes
associated with COVID-19 pathogenesis and the presence of viremia.
This study aims to investigate the association between SARS-CoV-2 viremia and various SNPs within genes that have previously been studied as predictors of COVID-19 severity.
Genetic Factors Associated with Viremia
The study involved a retrospective observational analysis of 340 COVID-19 patients admitted to the University Hospital La Princesa in Madrid between March 2020 and December 2021, all of whom had undergone at least one viremia determination. Positive viremia was defined as a viral load exceeding the quantifiable threshold of 20 copies/ml. A total of 38 SNPs were genotyped, and a multivariate logistic regression was performed to assess their association with viremia.
The study population had an average age of 64.5 years, with 60.9% of patients being male and 79.4% belonging to the white non-Hispanic ethnicity group. Notably, only 126 out of the 340 patients (37.1%) exhibited at least one instance of positive viremia. After adjusting for confounding variables, several genetic variants showed significant associations with the risk of viremia.
Among these genetic variants, the presence of minor alleles of rs2071746 (HMOX1), specifically the T/T genotype, was linked to a substantially higher risk of viremia (OR 9.9, p < 0.0001). Similarly, rs78958998 (associated with SERPING1 expression) exhibited an association with viremia risk, with the A/T genotype resulting in an OR of 2.3 (p = 0.04) and the T/T genotype yielding an OR of 12.9 (p < 0.0001). Another SNP, rs713400, which serves as an expression Quantitative Trait Locus (eQTL) for TMPRSS2, showed a potential association with viremia risk (C/T + T/T genotype OR 1.86, p = 0.10). Conversely, the minor alleles of rs11052877 (CD69) were associated with a decreased risk of viremia, with the A/G genotype showing an OR of 0.5 (p = 0.04) and the G/G genotype resulting in an OR of 0.3 (p = 0.01). Other genetic variants linked to a reduced viremia risk included rs2660 (OAS1) with an A/G genotype OR of 0.6 (p = 0.08), rs896 (VIPR1) with a T/T genotype OR of 0.4 (p = 0.02), and rs33980500 (TRAF3IP2) with a C/T + T/T genotype OR of 0.3 (p = 0.01).
After nearly three years of the COVID-19 pandemic, the disease continues to manifest with considerable heterogeneity, posing a challenge for healthcare professionals in predicting its course. Among potential biomarkers, the presence of SARS-CoV-2 viremia has emerged as a reliable indicator of disease severity.
While genome-wide analysis studies have been conducted to identify genetic variants associated with COVID-19 severity, this study is the first to investigate the relationship between specific genetic variants and the detection of viremia.
The results of this study suggest that only a limited number of genetic variants are associated with SARS-CoV-2 viremia. Among these variants, rs2071746 (HMOX1), rs78958998 (SERPING1), rs713400 (TMPRSS2), rs11052877 (CD69), rs33980500 (TRAF3IP2), rs2660 (OAS1), and rs896 (VIPR1) demonstrated potential roles in influencing viremia prevalence.
Importantly, the study rigorously adjusted for confounding variables and demonstrated that the association between these genetic variants and viremia remained significant, even when accounting for COVID-19 severity. This finding highlights the complex interplay between genetic, sociodemographic, therapeutic, and clinical factors in determining both viremia and disease severity. Additionally, the analysis revealed that age and sex were crucial variables in predicting viremia, emphasizing that multiple factors with relatively small contributions collectively shape an individual's genetic susceptibility to COVID-19 outcomes.
Among the genetic variants examined, rs713400 in TMPRSS2 attracted attention due to its role in viral entry into host cells. The study indicated that individuals carrying one copy of the T allele at rs713400 may be more likely to exhibit viremia. However, after adjusting for COVID-19 severity, the significance of this SNP diminished. This suggests that TMPRSS2 genetic variants may contribute to both viremia and disease severity, with alterations in TMPRSS2 expression potentially affecting the virus's ability to infect host cells and spread.
Another genetic variant of interest was rs33980500 in TRAF3IP2, associated with psoriasis and involved in immune regulation. The presence of the T allele in this SNP may lead to weaker activation of proinflammatory pathways dependent on IL-17, potentially resulting in better viral control and reduced viremia.
CD69, a gene involved in immune system regulation, exhibited an intriguing relationship with viremia. Patients carrying the minor allele of rs11052877 demonstrated a reduced risk of viremia, which may be attributed to increased CD69 expression. This heightened CD69 expression could potentially lead to decreased Th17 responses, resulting in less inflammatory reactions while maintaining effective viral control.
VIPR1, encoding the Vasoactive Intestinal Peptide receptor VPAC1, plays a role in anti-inflammatory responses. The study found that the T allele of rs896 was associated with a lower risk of viremia, possibly due to increased VPAC1 expression. Elevated VPAC1 expression could promote an anti-inflammatory response and inhibit Th17, contributing to reduced viremia.
OAS1, a key component of the interferon I pathway, is involved in controlling viral dissemination by degrading viral RNA. The study indicated that the A/G genotype of rs2660 in OAS1 trended towards protection against viremia, consistent with prior evidence of increased OAS1 activity associated with the G/G genotype.
HMOX1, which encodes heme oxygenase one (HO-1), a protein with anti-inflammatory effects, was also implicated in the study. Patients with the T allele of rs2071746 exhibited higher levels of viremia, potentially due to increased HO-1 expression, which could have anti-inflammatory and antiviral effects.
Lastly, rs78958998, an expression Quantitative Trait Locus (eQTL) for SERPING1, was linked to viremia risk. SERPING1 encodes the protein C1 inhibitor (C1INH), which is involved in regulating complement and coagulation pathways. Impaired SERPING1 expression could contribute to uncontrolled activation of complement and coagulation cascades, potentially promoting virus dissemination and viremia.
Despite these compelling associations, it's important to acknowledge that many of the identified SNPs have not been extensively studied in the context of COVID-19. Moreover, functional studies are needed to establish clear correlations between these genetic variants, gene expression, and protein activity.
The primary limitation of this study is its relatively small sample size, influenced by previous studies conducted by the research group. However, this sample size was sufficient to identify significant differences in the SNPs with the most pronounced effects. Expanding the scope of genetic variations and including a larger number of patients would be ideal but may be constrained by economic considerations and the need for larger cohorts. Additionally, the study lacked data on SARS-CoV-2 variants and vaccination status, which could potentially impact infectivity and viremia.
In conclusion, this study has demonstrated an association between specific genetic variants and the prevalence of SARS-CoV-2 viremia, even after adjusting for key clinical variables. The findings suggest that genetic factors, in combination with other demographic and clinical factors, contribute to the complex interplay influencing viremia and COVID-19 severity. While the implications of these identified variants in viremia prevalence are promising, further research and validation in diverse cohorts are necessary to establish their significance in the broader context of COVID-19.
The study findings were published in the peer reviewed journal: Frontiers in Medicine.
https://www.frontiersin.org/articles/10.3389/fmed.2023.1215246/full
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
