Study Discovers That SARS-CoV-2 ORF10 Causes Break Down Of Mitochondrial Antiviral Signaling Protein (MAVS), Suppressing Host Immune Response
Source: Medical News - ORF10 Dec 06, 2021 3 years, 4 months, 2 weeks, 8 hours, 41 minutes ago
A new study by researchers from Shandong Normal University-China and Changchun Veterinary Research Institute-China has discovered that the SARS-CoV-2 open reading frame 10 or ORF10 causes break down of mitochondrial antiviral signaling protein (MAVS), in the process suppressing the human host immune response.
To date, the function of open reading frame 10 (ORF10), which is uniquely expressed by SARS-CoV-2, remains unclear.
The study team demonstrated that overexpression of ORF10 markedly suppressed the expression of type I interferon (IFN-I) genes and IFN-stimulated genes.
The mitochondrial antiviral signaling protein (MAVS) was identified as the target via which ORF10 suppresses the IFN-I signaling pathway, and MAVS was found to be degraded through the ORF10-induced autophagy pathway.
It was also found that the overexpression of ORF10 promoted the accumulation of LC3 in mitochondria and induced mitophagy. Mechanistically, ORF10 was translocated to mitochondria by interacting with the mitophagy receptor Nip3-like protein X (NIX) and induced mitophagy through its interaction with both NIX and LC3B. Moreover, knockdown of NIX expression blocked mitophagy activation, MAVS degradation, and IFN-I signaling pathway inhibition by ORF10.
The study findings concluded that in the context of SARS-CoV-2 infection, ORF10 inhibited MAVS expression and facilitated viral replication. The findings reveal a novel mechanism by which SARS-CoV-2 inhibits the innate immune response; that is, ORF10 induces mitophagy-mediated MAVS degradation by binding to NIX.
The study findings were published in the peer reviewed journal: Cellular & Molecular Immunology (By Nature).
https://www.nature.com/articles/s41423-021-00807-4
To date, at least 26 ORFs have been identified in the SARS-CoV-2 genome but unfortunately many virologist and genomic specialist belong to the ‘old school' only believe that the five main ORFs (3a, 6, 7a, 7b, 8) are only the key canonical accessory proteins of significance.
https://pubmed.ncbi.nlm.nih.gov/32330414/
The only published study showing about 23 ORFs was done by researchers from Israel but a latest non published study shows 26 ORFs in the SARS-CoV-2 genome.
https://www.nature.com/articles/s41586-020-2739-1
Many of these ORFs and accessory proteins are believed to play a very important role in the SARS-CoV-2 pathogenesis and ways in affects the cellular systems and pathways of the human host but unfortunately not much attention is being paid to them.
There is a huge variety and diversity of coronaviruses (CoVs) that infect humans, pigs, cattle, horses, rats, and bats, among other host species. These viruses mainly cause infections of the respiratory and digestive tracts, with a wide range of clinical symptoms.
In the past, humans CoVs were thought to be relatively harmless respiratory infections. Zoonotic CoVs, on the other hand, have crossed species boundaries, resulting in novel CoVs that can transfer from animals to humans.
The SARS-CoV that caused SARS outbreaks in 2002 and 2003 and MERS-CoV that emerged in 2012 and continues to circulate in ca
mels are such examples as also the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19) that emerged in December 2019 and is responsible for the current severe COVID-19 outbreak worldwide, which has resulted in significant morbidity and mortality.
Corresponding author, Professor Dr Yuwei Gao from Changchun Veterinary Research Institute told Thailand
Medical News, “It is very critical and essential to have a deeper understanding of the molecular mechanisms behind viral infection and dissemination in the body along with the understanding of the various viral genes and proteins. The various ORFs and accessory proteins of the SARS-CoV-2 play a very critical role in pathogenesis and affccting the various cellular pathways of the human host.”
The SARS-CoV-2 genome is a positive-sense, nonsegmented, single-stranded RNA with a length of 29.9 kb. Many researchers unfortunately insist that it only has 14 open reading frames (ORFs) flanked by 5′ and 3′ untranslated regions in its genome and often only pay attention to the first few ORFs.
The ORF1a and ORF1b, which encode polyproteins that are subsequently cleaved to create 16 non-structural proteins (nsps) that make up the viral replicase-transcriptase complex, account for around two-thirds of the viral RNA genome. The spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, which are components of the virion, are encoded by ORFs in a third of the genome around the 3′ terminus.
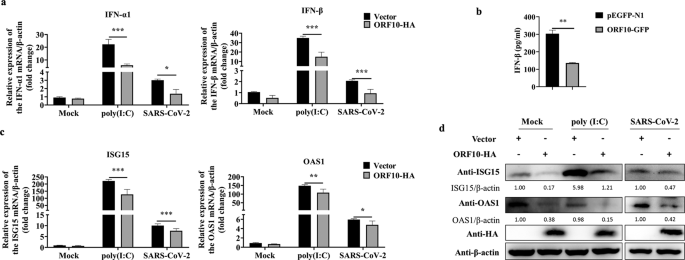 a HeLa-ACE2 cells were transfected with the ORF10-HA plasmid or empty vector (pCMV-HA) for 24 h and were then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1), and the transcription levels of IFN-α1 and IFN-β were monitored by qRT–PCR. b HeLa cells transfected with the ORF10-GFP plasmid or pEGFP-N1 were treated with poly(I:C) (20 μg/mL) for 24 h. The relative expression level of IFN-β in the supernatant was measured by ELISA. c qRT–PCR analysis of ISG15 and OAS1 mRNA expression in HeLa-ACE2 cells transfected with the ORF10-HA or pCMV-HA plasmid for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). d Immunoblot analysis of ISG15, OAS1, and β-actin expression in HeLa cells transfected with ORF10-HA or pCMV-HA for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). Data from three independent experiments were analyzed using t tests (a–c) and are presented as the mean ± SD values (*p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates a nonsignificant difference)
a HeLa-ACE2 cells were transfected with the ORF10-HA plasmid or empty vector (pCMV-HA) for 24 h and were then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1), and the transcription levels of IFN-α1 and IFN-β were monitored by qRT–PCR. b HeLa cells transfected with the ORF10-GFP plasmid or pEGFP-N1 were treated with poly(I:C) (20 μg/mL) for 24 h. The relative expression level of IFN-β in the supernatant was measured by ELISA. c qRT–PCR analysis of ISG15 and OAS1 mRNA expression in HeLa-ACE2 cells transfected with the ORF10-HA or pCMV-HA plasmid for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). d Immunoblot analysis of ISG15, OAS1, and β-actin expression in HeLa cells transfected with ORF10-HA or pCMV-HA for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). Data from three independent experiments were analyzed using t tests (a–c) and are presented as the mean ± SD values (*p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates a nonsignificant difference)
The new study findings show that the SARS-CoV-2 ORF10 reduces innate immunity and enhances viral replication by activating mitophagy to break down mitochondrial antiviral signaling protein (MAVS).
The study findings showed that ORF10 binds to Nip3-like protein X (NIX) and microtubule-associated protein 1 light chain 3 B (LC3B), then translocates to mitochondria, inducing mitophagy and MAVS destruction. NIX is also implicated in the ORF10-mediated degradation of MAVS and the inhibition of IFN responses
The study employs a number of techniques to prevent the generation of type I interferons (IFN-I,) allowing the virus to replicate quickly and avoid host antiviral responses. HeLa- angiotensin-converting enzyme 2 (ACE2) cells were transfected with ORF10-HA and an empty vector (pCMV-HA) to investigate the role of ORF10 in the antiviral innate immune response. Whole-cell lysates were then investigated using quantitative real-time PCR (qRT–PCR) and western blotting.
Interestingly when stimulated with poly(I:C) or infected with SARS-CoV-2, overexpression of ORF10 significantly reduced transcription of the IFN-1 and IFN- genes.
The study team also discovered that overexpression of ORF10 inhibited the production of IFN- in HeLa cells when stimulated with poly(I:C) (I:C). Following that, the effect of ORF10 expression on antiviral ISGs was investigated, and it was discovered that after poly(I:C) treatment or SARS-CoV-2 infection, ORF10 expression lowered the mRNA levels of ISG15 and OAS1 relative to control cells.
Typically, mitochondrial MAVS connects antiviral type I IFN signaling to mitochondria. Some viruses cause mitophagy, which results in mitochondrial fragmentation and, as a result, a reduction in innate immunity.
Significantly, in SARS-CoV-2-infected cells, green fluorescent protein- (GFP-) tagged LC3 colocalized with DsRed-tagged Mito, a mitochondrial marker protein, showing that SARS-CoV-2 infection triggered mitophagy.
Subsequent treatment with bafilomycin A1 (BafA1) or mitochondrial division inhibitor 1 (Mdivi-1) prevented MAVS degradation generated by SARS-CoV-2 infection, implying that SARS-CoV-2 infection causes MAVS degradation via mitophagy.
In order to see if ORF10 inhibits MAVS protein expression by triggering mitophagy, the authors looked at where ORF10 is found in mitochondria.
In ORF10-GFP-transfected HeLa cells, immunofluorescence analysis revealed that GFP-tagged ORF10 colocalized with DsRed-tagged Mito.
The subsequent step was to see if ORF10 could trigger mitophagy. In ORF10-HA-transfected HeLa cells, immunofluorescence examination revealed that GFP-tagged LC3B colocalized with DsRed-tagged Mito, showing that ORF10 caused LC3 localization in mitochondria.
Also, the study team detected the expression of the outer membrane mitochondrial protein TOMM20, a mitochondrial marker protein. They discovered that ORF10 expression decreased translocase of outer mitochondrial membrane 20 (TOMM20) expression, which was prevented by Baf A1 treatment. SARS-CoV-2 ORF10 destroys the MAVS protein via mitophagy, according to these study findings.
The study findings conclude that the SARS-CoV-2 ORF10 protein suppresses innate antiviral defense and promotes viral replication. These data suggest that ORF10 interacts with NIX and LC3B to trigger mitophagy, resulting in MAVS-mediated antiviral signaling being blocked.
The study findings of this work open up new avenues for research into the molecular mechanisms by which SARS-CoV-2 evades innate antiviral immunity.
Please help to sustain this site and also all our research and community initiatives by making a donation. Your help means a lot and helps saves lives directly and indirectly and we desperately also need financial help now.
https://www.thailandmedical.news/
For the latest
SARS-CoV-2 Research, keep on logging to Thailand Medical News.

 a HeLa-ACE2 cells were transfected with the ORF10-HA plasmid or empty vector (pCMV-HA) for 24 h and were then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1), and the transcription levels of IFN-α1 and IFN-β were monitored by qRT–PCR. b HeLa cells transfected with the ORF10-GFP plasmid or pEGFP-N1 were treated with poly(I:C) (20 μg/mL) for 24 h. The relative expression level of IFN-β in the supernatant was measured by ELISA. c qRT–PCR analysis of ISG15 and OAS1 mRNA expression in HeLa-ACE2 cells transfected with the ORF10-HA or pCMV-HA plasmid for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). d Immunoblot analysis of ISG15, OAS1, and β-actin expression in HeLa cells transfected with ORF10-HA or pCMV-HA for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). Data from three independent experiments were analyzed using t tests (a–c) and are presented as the mean ± SD values (*p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates a nonsignificant difference)
a HeLa-ACE2 cells were transfected with the ORF10-HA plasmid or empty vector (pCMV-HA) for 24 h and were then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1), and the transcription levels of IFN-α1 and IFN-β were monitored by qRT–PCR. b HeLa cells transfected with the ORF10-GFP plasmid or pEGFP-N1 were treated with poly(I:C) (20 μg/mL) for 24 h. The relative expression level of IFN-β in the supernatant was measured by ELISA. c qRT–PCR analysis of ISG15 and OAS1 mRNA expression in HeLa-ACE2 cells transfected with the ORF10-HA or pCMV-HA plasmid for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). d Immunoblot analysis of ISG15, OAS1, and β-actin expression in HeLa cells transfected with ORF10-HA or pCMV-HA for 24 h and then treated with poly(I:C) (20 μg/mL) or infected with SARS-CoV-2 (MOI = 0.1). Data from three independent experiments were analyzed using t tests (a–c) and are presented as the mean ± SD values (*p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates a nonsignificant difference)