Study Reveals That Noncoding Regions Of The SARS-CoV-2 RNA (ncrRNAs) Play Key Role In The Virus-Host Interactions
COVID-19 News - Noncoding Regions Of The SARS-CoV-2 RNA (ncrRNAs) Jun 15, 2023 1 year, 10 months, 5 days, 14 hours, 1 minute ago
High-Sensitivity Profiling Of SARS-CoV-2 Noncoding Region–Host Protein Interactome Reveals The Potential Regulatory Role Of Negative-Sense Viral RNA
COVID-19 News: A new study conducted at Zhejiang University in Hangzhou, China, has revealed a fascinating aspect of the SARS-CoV-2 virus that could revolutionize our understanding of its behavior. The study focuses on the noncoding regions of the viral RNA, known as ncrRNAs, and their pivotal role in virus-host interactions. These findings have the potential to reshape our approach to therapeutics and pave the way for tackling emerging coronaviruses.
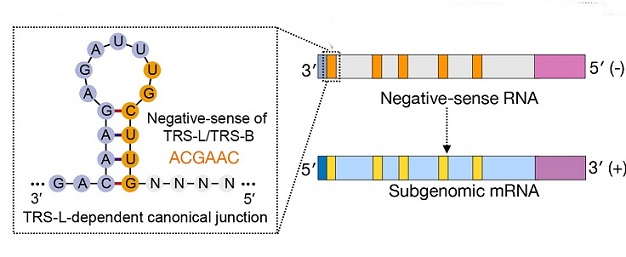
In order to comprehensively examine the ncrRNA-host protein interactome, the study team developed an innovative method that combined MS2 affinity purification with liquid chromatography-mass spectrometry. They also designed a diverse set of bait ncrRNAs to systematically map the interactions between SARS-CoV-2 ncrRNA and cells such as Calu-3, Huh7, and HEK293T.
The integration of the results allowed the researchers to identify the core ncrRNA-host protein interactomes across different cell lines. They discovered that the 5' untranslated region (UTR) interactome is rich in proteins from the small nuclear ribonucleoproteins family, which play a critical role in regulating viral replication and transcription. On the other hand, the 3' UTR interactome is enriched with proteins involved in stress granules and the heterogeneous nuclear ribonucleoproteins family.
What's particularly intriguing is the extensive range of host proteins that the negative-sense ncrRNAs, especially the negative-sense 3' UTR, interacted with across all cell lines. These proteins are key players in the regulation of viral production processes, host cell apoptosis, and immune response. This discovery sheds new light on the vital role that negative-sense ncrRNAs play during infection and opens up exciting possibilities for designing future therapeutics. The findings suggest that the negative-sense ncrRNAs play a vital role in manipulating the host cellular machinery to facilitate viral replication and evade the immune system.
The study's findings have far-reaching implications, extending beyond SARS-CoV-2. The highly conserved nature of untranslated regions in positive-strand viruses suggests that the regulatory role of negative-sense ncrRNAs may not be exclusive to SARS-CoV-2 alone.
Understanding the intricate interactions between viral RNAs and host proteins is crucial for unraveling the biology of SARS-CoV-2 and devising effective strategies against COVID-19. The researchers' innovative MS2 affinity purification and liquid chromatography-mass spectrometry method, known as MAMS, allowed them to systematically map the viral ncrRNA-host protein interactome across multiple cell lines with exceptional sensitivity.
Of particular interest is the interaction between the viral 5' UTR and proteins involved in U1 small nuclear ribonucleoprotein (snRNP). These interactions could potentially serve as targets for antiviral drugs. Additionally, the study identified antiviral factors, such as ZC3HAV1, MOV10, and EIF2AK2, that interacted with the 5' UTR, inhibiting viral replication and transcription.
The viral 3' UTR, on the othe
r hand, exhibited strong associations with proteins related to stress granules (SGs) and the heterogeneous nuclear ribonucleoprotein (hnRNP) family. SARS-CoV-2 exploits these interactions to inhibit the formation of antiviral SGs, thereby maximizing viral replication efficiency. Understanding the interplay between the 3' UTR and SGs can provide valuable insights into the transcription and replication processes of SARS-CoV-2.
The study also highlighted the intriguing potential of negative-sense ncrRNAs in influencing viral infection. Negative-sense RNAs are traditionally considered as templates for synthesizing positive-sense genomic RNA and subgenomic RNAs in positive-strand RNA viruses. However, this research revealed that negative-sense ncrRNAs of SARS-CoV-2 interacted with a wide range of host proteins, indicating that they may have important regulatory roles in various stages of viral infection.
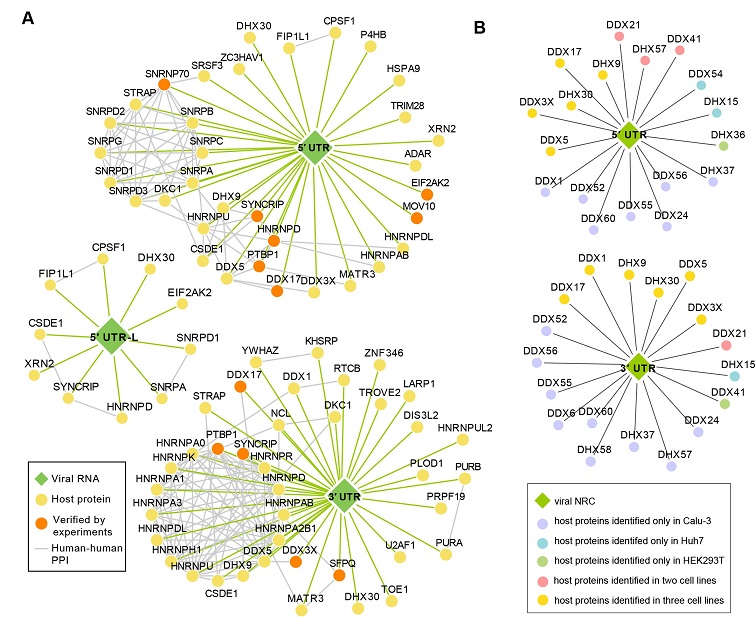 The core SARS-CoV-2 ncrRNA interactome. (A) Core interactome of SARS-CoV-2 ncrRNAs. Each node represents a host protein (closed circle) or viral ncrRNA (diamond). Orange circles represent that the interactions between host proteins and viral ncrRNA were verified in the RIP experiments. Green edges indicate the interactions between viral ncrRNAs and host proteins. Gray edges denote the interactions in host proteins. (B) Network revealing the interactions between DEAD/DEAH-box helicases and 5′ UTR/3′ UTR. Each node represents a host protein (closed circle) or viral ncrRNA (diamond). Colors are denoted in the box.
The core SARS-CoV-2 ncrRNA interactome. (A) Core interactome of SARS-CoV-2 ncrRNAs. Each node represents a host protein (closed circle) or viral ncrRNA (diamond). Orange circles represent that the interactions between host proteins and viral ncrRNA were verified in the RIP experiments. Green edges indicate the interactions between viral ncrRNAs and host proteins. Gray edges denote the interactions in host proteins. (B) Network revealing the interactions between DEAD/DEAH-box helicases and 5′ UTR/3′ UTR. Each node represents a host protein (closed circle) or viral ncrRNA (diamond). Colors are denoted in the box.
The study also examined the enigmatic role of ORF10, a gene in the SARS-CoV-2 genome whose function remains uncertain and has never been covered in detail in any previous studies or
COVID-19 News reports. Previous genome annotations positioned ORF10 upstream of the 3' UTR, but evidence suggests that it may not encode a functional protein. Instead, the negative-sense ncrRNAs derived from ORF10, along with other distinct negative-sense ncrRNAs, demonstrated interactions with proteins involved in viral processes, mitochondrial translation, ERAD pathway, and immune response. These findings provide intriguing insights into the potential functional roles of negative-sense ncrRNAs, highlighting their significance in SARS-CoV-2 infection.
The implications of this study extend beyond SARS-CoV-2. The highly conserved nature of untranslated regions (UTRs) in positive-strand RNA viruses, including coronaviruses and more distantly related viruses like Zika and Dengue, suggests that negative-sense ncrRNAs in these viruses may also possess regulatory roles in virus-host interactions. Therefore, further research into the regulatory mechanisms of negative-sense ncrRNAs could potentially unlock new avenues for antiviral drug discovery against a range of viral infections.
These new study findings provide a comprehensive landscape of the SARS-CoV-2 ncrRNA-host protein interactome, uncovering the crucial role of negative-sense ncrRNAs in virus-host interactions. The findings not only deepen our understanding of the molecular mechanisms underlying SARS-CoV-2 infection but also pave the way for the development of novel therapeutic strategies targeting the viral ncrRNA-host protein interactions. With the ongoing threat of emerging coronaviruses and the need for effective antiviral treatments, this study marks a significant step forward in our battle against the COVID-19 diseases.
The study findings were published in the peer reviewed journal: mSystems.
https://journals.asm.org/doi/10.1128/msystems.00135-23
For the latest
COVID-19 News, keep on logging to Thailand Medical News.

