Swiss Study Uncovers That HLA Variants And T-Cell Receptor (TCR) Diversity Play A Role In COVID-19 Infection Outcomes!
COVID-19 News - HLA Variants - T-cell Receptor (TCR) Diversity Jul 19, 2023 2 years, 6 months, 2 weeks, 2 days, 18 hours ago
Heterologous immunity and the random somatic recombination of TCRs may underpin the efficacy of the immune response against SARS-CoV-2 in individuals without immunodeficiency risk factors, particularly those who were infected prior to vaccination.
COVID-19 News: Acute infection caused by the SARS-CoV-2 coronavirus has swept across the globe, bringing forth a myriad of outcomes in infected individuals. As scientists race to understand the factors contributing to COVID-19 susceptibility and severity, the crucial role of HLA antigen presentation and T-cell mediated immunity is being unraveled.
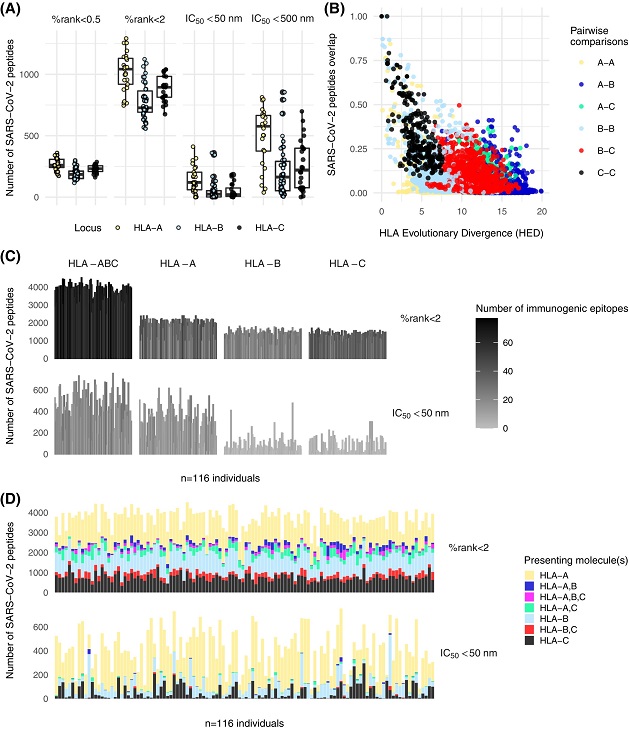 SARS-CoV-2 peptide binding predictions. (A) Number of peptides presented by HLA class I alleles sorted by locus, using both prediction scores at two binding thresholds. (B) Relationship between peptide overlap estimated from strong and weak binders and HLA evolutionary divergence (HED). (C) Size of the HLA class I peptide repertoire in 116 recipients for peptides considered as strong and weak binders according to percentile rank score and strong binders according to affinity score. The data are presented for combined loci and at the single locus level. The black and white gradient indicates the number of immunogenic epitopes for each recipient among a set of 92 previously published HLA class I T-cell epitopes. (D) Same as (C), but only for the combined loci and by distinguishing the presenting HLA molecule(s).
SARS-CoV-2 peptide binding predictions. (A) Number of peptides presented by HLA class I alleles sorted by locus, using both prediction scores at two binding thresholds. (B) Relationship between peptide overlap estimated from strong and weak binders and HLA evolutionary divergence (HED). (C) Size of the HLA class I peptide repertoire in 116 recipients for peptides considered as strong and weak binders according to percentile rank score and strong binders according to affinity score. The data are presented for combined loci and at the single locus level. The black and white gradient indicates the number of immunogenic epitopes for each recipient among a set of 92 previously published HLA class I T-cell epitopes. (D) Same as (C), but only for the combined loci and by distinguishing the presenting HLA molecule(s).
Recent research has shed light on the depth of peptide presentation and the breadth of the T-cell repertoire, both of which have been linked to disease outcomes.
Intriguingly, evidence has emerged that unexposed individuals can develop robust T-cell responses against SARS-CoV-2 due to the phenomenon of heterologous immunity.
In this groundbreaking study, researchers at Geneva University Hospitals delve into the intricate world of T-cell receptors (TCRs) and explore the anti-SARS-CoV-2 T-cell repertoire in the pre-COVID-19 era. By analyzing previously published TCR CDR3β immunosequencing data from a cohort of healthy donors and recipients of allogeneic hematopoietic stem cell transplantation (alloHSCT), they unveil fascinating insights into the genetic capacity to recognize and combat the SARS-CoV-2 virus.
A Genetic Arsenal
The researchers embarked on a comprehensive investigation, examining a staggering 143,310 publicly available SARS-CoV-2 specific T-cell sequences within a cohort of 116 individuals. Simultaneously, they performed HLA class I peptide binding predictions using the virus's reference proteome and high-resolution genotyping data.
Astonishingly, their findings demonstrated that every individual possessed the genetic weaponry required to recognize and respond to SARS-CoV-2 effectively.
This was evidenced by the median cumulative frequency of clonotypes with sequences matching SARS-CoV-2 specific T-cells, amounting to an impressive 5% within the repertoire.
Furthermore, regardless of the HLA class I variants present, individuals exhibited a broad capacity to present hundreds of SARS-CoV-2 derived peptides. These remarkable observations suggest that heterologous
immunity and the random somatic recombination of TCRs may underpin the efficacy of the immune response against SARS-CoV-2 in individuals without immunodeficiency risk factors, particularly those who were infected prior to vaccination.
Unveiling the T-Cell Repertoire
The T-cell repertoire is a dynamic and diverse landscape, orchestrating the body's immune defenses. To shed light on the SARS-CoV-2 specific T-cell landscape, the study team examined a cohort of 116 individuals, including both healthy donors and recipients of alloHSCT. They employed high-resolution HLA genotyping and surveyed the TCR CDR3β repertoire through immunosequencing.
Intriguingly, they discovered that each individual harbored a vast array of T cells with CDR3β sequences similar to those of SARS-CoV-2 specific clonotypes. Surprisingly, this phenomenon was not exclusive to SARS-CoV-2 but extended to other viruses such as HIV-1.
While the repertoire's clonality was slightly reduced in alloHSCT recipients compared to healthy donors, the overall presence of diverse clonotypes capable of recognizing the virus remained evident. These findings provide novel insights into the generation and composition of the T-cell repertoire, emphasizing the role of heterologous immunity and random somatic gene recombination in shaping a robust defense against viral infections.
Unveiling the Antigenic Presentation
To complement their analysis of the TCR repertoire, the study team delved into the capacity of patients to present SARS-CoV-2 antigenic peptides on their HLA class I molecules for T-cell recognition. They observed a wide range of binders, varying in number based on the HLA molecule and binding level considered.
Interestingly, HLA-A molecules exhibited a greater promiscuity in binding SARS-CoV-2 peptides, while HLA-B molecules displayed more selective preferences.
The study team also noted a correlation between the overlap in peptides presented by HLA class I molecules and their binding capacity. Each patient was predicted to present a substantial number of both strong and weak binders, with HLA-A molecules playing a prominent role. The heterogeneous profile of peptide presentation suggests that functional differences in antigen presentation may be associated with COVID-19 outcomes. Moreover, the presence of shared or similar antigenic peptides between SARS-CoV-2 and other seasonal coronaviruses underscores the potential role of heterologous immunity in shaping immune responses.
Implications for Immune Protection
The study's findings shed light on the complexity of interactions between TCRs and pHLAs, emphasizing the importance of redundancy and cross-reactivity as covered in previous studies and
COVID-19 News reports.
While each individual possesses a diverse repertoire of T cells capable of recognizing a wide range of SARS-CoV-2 peptides across various HLA backgrounds, subtle differences in the TCR repertoire and peptide presentation may exist.
While the genetic factors of HLA and TCR alone are insufficient to clear the virus, their presence suggests a critical foundation for an effective immune response when appropriately activated. However, it is crucial to acknowledge that individuals with compromised immune systems, such as alloHSCT recipients or those with underlying conditions, may face challenges in mounting an adequate immune response. These insights into the adaptative immune response contribute to our understanding of the mechanisms that drive susceptibility, severity, and clearance of SARS-CoV-2 infection.
Conclusion
The study findings offer an intriguing glimpse into the genetic factors underlying T-cell mediated immunity against SARS-CoV-2. The study reveals a vast repertoire of T cells capable of recognizing SARS-CoV-2-derived peptides in most HLA backgrounds, reflecting the potential for a robust immune response. However, the study also highlights the importance of a functional immune system in unleashing the full potential of these genetic factors. As the global fight against COVID-19 continues, these findings contribute to our understanding of the complex interplay between genetics, immunity, and viral infections, paving the way for future research and therapeutic interventions.
The study findings were published in the peer reviewed journal: HLA Immune Response genetics (Wiley)
https://onlinelibrary.wiley.com/doi/10.1111/tan.15158
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
