Nikhil Prasad Fact checked by:Thailand Medical News Team Dec 13, 2024 4 months, 2 days, 3 hours, 59 minutes ago
Medical News: Scientists from Goethe University in Frankfurt, Columbia University in New York, Deutsches Elektronen-Synchrotron in Hamburg, and the University of Greifswald have unraveled critical insights into the SARS-CoV-2 nucleocapsid protein. This protein, essential for the virus’ RNA genome processing, plays a pivotal role in the virus’ ability to survive, replicate, and spread. The research, conducted using advanced techniques such as nuclear magnetic resonance (NMR) spectroscopy and crystallography, sheds light on the mutations within the nucleocapsid’s N-terminal domain (NTD) and their effects on RNA-binding efficiency.
 The Core Network in SARS-CoV-2 and Its Role in RNA Binding
Why This Study Matters
The Core Network in SARS-CoV-2 and Its Role in RNA Binding
Why This Study Matters
Despite global vaccination efforts and reduced severity of COVID-19 cases, the virus continues to mutate. This
Medical News report focuses on a recent study exploring how structural mutations in the SARS-CoV-2 nucleocapsid protein impact the virus’ core mechanisms. Such studies are crucial as mutations in the virus’ genome pose potential risks, including increased infectivity or resistance to immune responses.
Overview of the SARS-CoV-2 Nucleocapsid Protein
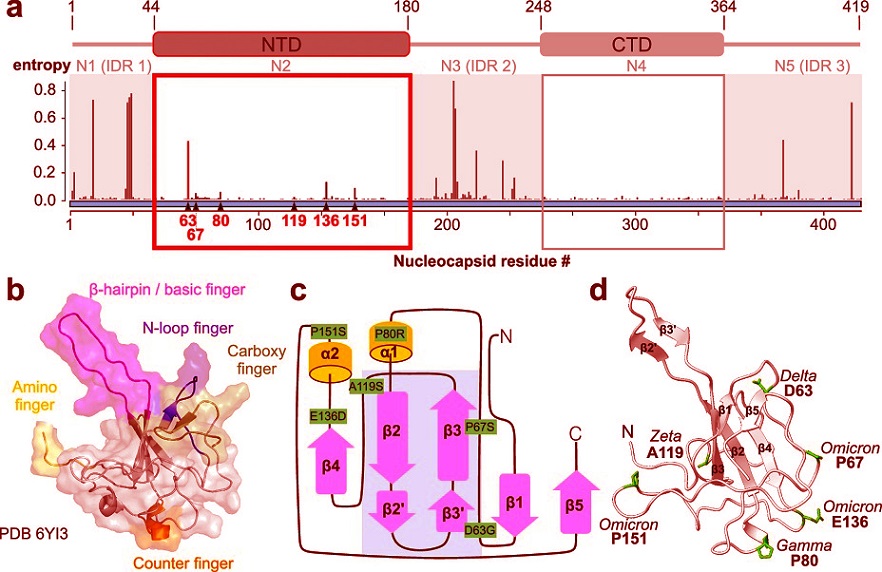
The nucleocapsid protein (N protein) of SARS-CoV-2 has multiple functions, ranging from assisting RNA replication to forming regulatory complexes. The N protein comprises several domains, but the NTD is particularly interesting due to its hand-like structure that allows it to bind RNA selectively. Previous studies have highlighted that this domain uses a specific core network to maintain its stability and flexibility, enabling the recognition and binding of viral RNA.
This research aimed to understand how naturally occurring mutations affect this core network and, in turn, the protein’s ability to function efficiently.
Key Findings of the Study
-Conservation of the NTD Structure: The NTD’s three-dimensional structure, which includes a “palm” and “fingers,” is essential for its role in RNA binding. The study found that this structure is preserved even in mutants with slight variations. This conservation indicates that the NTD’s stability is evolutionarily critical.
-The Core Network: At the center of the NTD lies a network of three residues - Q58, W108, and F171 - that anchor the structural integrity of the domain. Mutations disrupting this network significantly destabilize the protein, impairing its ability to bind RNA.
-Effect of Specific Mutations:
D63G and P80R Mutations: These mutants exhibited a slight increase in RNA-binding affinity compared to the wild type. This increased affinity might enhance the virus’ replication efficiency under certain conditions.
Q58I Mutation: This mutation disrupts the core ne
twork, leading to a significant loss of structural stability and specific RNA recognition capabilities.
W108G and F171G Mutations: These mutations destabilized the entire protein structure, confirming the vital role of these residues in maintaining the NTD’s function.
-RNA-Binding Specificity: The NTD’s ability to differentiate between viral RNA and other RNA molecules is dependent on the intact coordination between its palm and flexible fingers. Mutations that interfere with this coordination reduce the protein’s specificity, which could influence the virus’ overall fitness.
Implications of the Findings
The study emphasizes that even minor mutations can influence the functionality of the SARS-CoV-2 nucleocapsid protein. For instance, while some mutations increase RNA-binding affinity, they might also compromise the protein’s specificity, leading to potential inefficiencies in viral genome processing. Conversely, mutations that disrupt the core network could weaken the virus, providing a potential target for antiviral therapies.
Applications for Therapeutic Development
Understanding the role of the core network in the NTD opens avenues for drug development. Researchers can now target this network with small molecules to inhibit the nucleocapsid protein’s function. This approach could complement existing therapies by reducing the virus’ ability to replicate and spread.
Broader Evolutionary Insights
Interestingly, the core network’s conservation across different coronaviruses highlights its evolutionary importance. Betacoronaviruses, including SARS-CoV and MERS-CoV, share similar structural networks in their nucleocapsid proteins, suggesting that targeting this region could yield broad-spectrum antiviral strategies.
Conclusion
This study provides critical insights into how the SARS-CoV-2 nucleocapsid protein maintains its structural and functional integrity. By identifying a conserved core network essential for RNA binding, the research highlights potential vulnerabilities in the virus’ life cycle. Future therapies could exploit these vulnerabilities to develop more effective treatments for COVID-19 and related diseases.
The findings underscore the need for continued surveillance of viral mutations and their effects on protein function. As SARS-CoV-2 continues to evolve, understanding its molecular mechanisms will remain crucial in managing the pandemic and preparing for future outbreaks.
The study findings were published in the peer-reviewed journal: Nature Communications.
https://link.springer.com/article/10.1038/s41467-024-55024-0
For the latest COVID-19 News, keep on logging to Thailand
Medical News.
Read Also:
https://www.thailandmedical.news/news/the-role-of-g3bp-in-sars-cov-2-replication
https://www.thailandmedical.news/news/new-mutation-emerges-in-sars-cov-2-nsp13-protein-that-allows-escape-from-nk-cell-recognition
https://www.thailandmedical.news/articles/coronavirus
Follow Us on:
https://x.com/ThailandMedicaX
https://bsky.app/profile/thailandmedical.bsky.social
https://www.facebook.com/ThailandMedicalNews
https://gettr.com/user/thailandmedicalnews
https://www.tribel.com/thailandmedical/wall
