Unveiling the Secrets of The New SARS-CoV-2 Omicron Sublineages: Dynamic Signatures, Conformational Changes and Hidden Allosteric Pockets Revealed
COVID-19 News - Hidden Allosteric Pockets In New Omicron Sublineages May 25, 2023 1 year, 10 months, 3 weeks, 5 days, 11 hours, 40 minutes ago
The Intriguing Interplay of Dynamics and Convergent Evolution Modulates Allostery and Functional Mechanisms
COVID-19 News: In a stunning revelation, researchers from Southern Methodist University in Dallas, Texas, and Chapman University in Orange, California, have uncovered groundbreaking insights into the dynamic behavior and hidden secrets of the emerging SARS-CoV-2 Omicron variants. These variants, including XBB.1, XBB.1.5, BQ.1, and BQ.1.1, have gained significant attention due to their increased transmissibility and potential immune evasion capabilities. By combining cutting-edge computational techniques and extensive molecular simulations, the scientists have delved deep into the intricate interplay between the viral spike proteins and the host receptor ACE2, unraveling distinct dynamic signatures and hidden allosteric pockets that govern the virus's behavior.
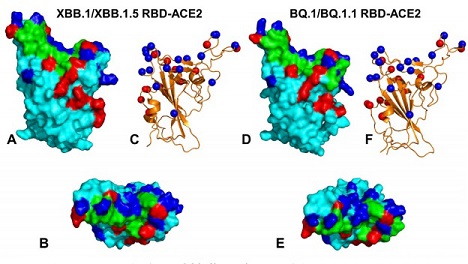 Structural organization & binding epitopes of the SARS-CoV-2-RBD Omicron XBB.1/XBB.1.5 and BQ.1/BQ.1.1 complexes with human ACE enzyme. (A) The optimized modeled structure of the RBD XBB.1/XBB.1.5-ACE2 complex.The RBD is in cyan surface, The binding epitope is in green surface, XBB.1/XBB.1.5 Omicron mutations are in red, and the convergent mutational sites are in blue surface. (B) The top view of the RBD XBB.1/XBB.1.5 from complexes with ACE2. (C) The Omicron RBD XBB.1/XBB.1.5 in ribbon representation. The sites of convergent mutations are in blue spheres. (D) The optimized modeled structure of the RBD BQ.1/BQ.1.1-ACE2 complex. The RBD is in cyan surface, The binding epitope is in green surface, BQ.1/BQ.1.1 Omicron mutations are in red, and the convergent mutational sites (R346T, K356, N440K, K444T, V445, G446, N450, L452R, N460K, T478K, E484A, F486V, F490, Q493, S494, N501Y) are in blue surface. (E) The top view of the RBD BQ.1/BQ.1.1 from complexes with ACE2. (F) The RBD BQ.1/BQ.1.1 in ribbon representation. The sites of convergent mutations are shown in blue spheres
The Era of Convergent Evolution
Structural organization & binding epitopes of the SARS-CoV-2-RBD Omicron XBB.1/XBB.1.5 and BQ.1/BQ.1.1 complexes with human ACE enzyme. (A) The optimized modeled structure of the RBD XBB.1/XBB.1.5-ACE2 complex.The RBD is in cyan surface, The binding epitope is in green surface, XBB.1/XBB.1.5 Omicron mutations are in red, and the convergent mutational sites are in blue surface. (B) The top view of the RBD XBB.1/XBB.1.5 from complexes with ACE2. (C) The Omicron RBD XBB.1/XBB.1.5 in ribbon representation. The sites of convergent mutations are in blue spheres. (D) The optimized modeled structure of the RBD BQ.1/BQ.1.1-ACE2 complex. The RBD is in cyan surface, The binding epitope is in green surface, BQ.1/BQ.1.1 Omicron mutations are in red, and the convergent mutational sites (R346T, K356, N440K, K444T, V445, G446, N450, L452R, N460K, T478K, E484A, F486V, F490, Q493, S494, N501Y) are in blue surface. (E) The top view of the RBD BQ.1/BQ.1.1 from complexes with ACE2. (F) The RBD BQ.1/BQ.1.1 in ribbon representation. The sites of convergent mutations are shown in blue spheres
The Era of Convergent Evolution
The relentless evolutionary pressure on the SARS-CoV-2 virus has led to the emergence of highly transmissible Omicron variants. These variants, through a process known as convergent evolution, have acquired multiple mutations that provide them with a significant growth advantage and enhanced viral fitness as shown in previous studies and covered in previous
COVID-19 News reports. This phenomenon suggests that the immune pressure exerted by host immune responses can inadvertently promote the rapid evolution of the virus. The researchers meticulously studied the conformational landscapes of these Omicron variants, employing state-of-the-art structural modeling, extensive microsecond molecular dynamics (MD) simulations, and Markov state models.
Unveiling Dynamic Signatures
Through their comprehensive simulations, the study team shed light on the conformational dynamics of the SARS-CoV-2 spike complexes with the ACE2 receptor for the XBB.1, XBB.1.5, BQ.1, and BQ.1.1 Omicron variants.
Remarkably, despite their structural sim
ilarities, these variants exhibited unique dynamic signatures and distinct distributions of conformational states. The findings suggested that variant-specific changes in the functional interfacial loops of the spike receptor binding domain can be finely tuned through cross-talk between convergent mutations, thereby facilitating immune escape and modulation of viral infectivity. The researchers' integrative approach provided a detailed understanding of the interplay between dynamics and convergent evolution, offering insights into the underlying mechanisms of viral evolution and adaptation.
Revealing Hidden Allosteric Pockets
Allostery, the phenomenon of long-range communication within proteins, plays a crucial role in modulating their functions. The study team employed perturbation-based approaches and combined them with atomistic simulations and Markovian modeling analysis to uncover the intricate allosteric networks at play in the SARS-CoV-2 Omicron complexes. This integrative computational approach revealed the existence of hidden allosteric pockets within the spike proteins, further enhancing our understanding of their functional mechanisms. The study team demonstrated that convergent mutation sites act as both effectors and receivers of allosteric signaling, regulating conformational plasticity at the binding interface and modulating the virus's allosteric responses.
The Dance of Dynamics and Allostery
The interplay between dynamics and allostery emerged as a central theme in this groundbreaking study. The study team demonstrated how the intricate dance of conformational changes, driven by dynamic signatures and allosteric networks, governs the behavior of the SARS-CoV-2 Omicron variants.
Markov modeling and microsecond simulations and provided a detailed characterization of the conformational landscapes and revealed the increased thermodynamic stabilization of the XBB.1.5 subvariant which is contrasted to more dynamic BQ.1 and BQ.1.1 subvariants. Despite considerable structural similarities, Omicron mutations can induce unique dynamic signatures and specific distributions of conformational states that explain the patterns of stability and immune escape in the Omicron variants.
Utilizing a comparative MSM analysis we showed that the distribution of dominant macrostates in the Omicron variants can allow for allosteric modulation of mobility in the flexible RBD loops 470-491 and 440-452 involved in the interfacial contacts and containing critical convergent mutation sites.
The findings suggested that variant-specific changes of conformational mobility in these functional loops can be fine-tuned through cross-talk between convergent mutations thereby providing an evolutionary path for the improved immune escape without significant cost on the ACE2 binding.
By combining MD simulations and MSM analysis with perturbation-based response scanning approaches, the study team also examined mechanisms of long-range dynamic couplings and allosteric communications in the Omicron RBD-ACE2 complexes. Based on premise that allostery is linked with the evolvability of proteins, they explored the relationship between allosteric interactions and patterns of convergent Omicron mutations, showing how convergent mutations can potentiate conformational plasticity and modulate allosteric responses in the RBD to binding and immune escape.
Through perturbation response scanning analysis, the study reveals important complementary roles of convergent mutation sites as effectors/regulators and sensors/receivers of allosteric signal transmission in the RBDACE2 complexes.
The study team also explored the reversed allosteric communication approach and characterized the effect of dynamics on the distribution of allosteric pockets in the Omicron subvariants. The results show that variant-specific redistribution of macrostates preserves the experimentally known allosteric pocket on the RBD while allowing for the emergence of hidden allosteric pockets that are anchored by convergent mutation sites K444T and L452R. The findings support a mechanism according to which Omicron mutations may have evolved to balance thermodynamic stability and conformational adaptability in order to ensure proper tradeoff between stability, binding and immune escape.
The study findings highlighted the role of specific mutations in fine-tuning the virus's ability to interact with the ACE2 receptor and evade the host immune response. By deciphering the secrets of dynamic signatures and hidden allosteric pockets, this research opens new avenues for designing targeted therapeutics and vaccines against the various new Omicron sub-lineages and recombinant sub-lineages.
Implications and Further Research
The understanding of the dynamic signatures and allosteric networks involved in the behavior of the Omicron variant provides valuable insights for developing effective strategies to combat the virus. With this knowledge, scientists can explore the design of targeted therapeutics that specifically disrupt the allosteric communication pathways critical for viral entry and replication.
One potential approach is the development of small molecules or peptides that can bind to the allosteric pockets identified in the study. By binding to these pockets, these molecules can modulate the conformational dynamics of the viral proteins, inhibiting their function and preventing viral replication. Targeting allosteric sites can offer advantages over traditional approaches that focus solely on inhibiting the active site of a protein, as allosteric sites often exhibit higher specificity and lower chances of resistance development.
Furthermore, the insights gained from this research can aid in the development of vaccines tailored to combat the Omicron variant. Vaccines typically work by eliciting an immune response that recognizes and neutralizes specific viral proteins. By understanding the dynamic changes and allosteric interactions that influence the viral proteins' behavior, scientists can design vaccines that target these critical regions. This approach could enhance the immune response's effectiveness, ensuring a robust defense against the Omicron variant.
In addition to therapeutic and vaccine development, the study's findings also pave the way for further investigations into other emerging variants or viruses. The dance of dynamics and allostery is likely to be a universal principle governing the behavior of many pathogens. By applying similar methodologies and approaches, scientists can gain a deeper understanding of other viral systems and develop broader strategies to combat future viral threats.
Overall, the research on the interplay between dynamics and allostery in the context of the SARS-CoV-2 Omicron variants offers a promising avenue for the development of targeted therapeutics, vaccines, and a more comprehensive understanding of viral behavior. By unraveling the secrets of how viruses exploit conformational changes and allosteric networks, scientists can work towards mitigating the impact of viral infections and protecting global health.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2023.05.20.541592v1
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
